Stromal endothelial cells establish a bidirectional crosstalk with chronic lymphocytic leukemia cells through the TNF-related factors BAFF, APRIL, and CD40L
- PMID: 22593611
- PMCID: PMC3370079
- DOI: 10.4049/jimmunol.1102066
Stromal endothelial cells establish a bidirectional crosstalk with chronic lymphocytic leukemia cells through the TNF-related factors BAFF, APRIL, and CD40L
Abstract
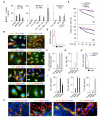
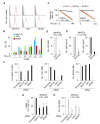
Chronic lymphocytic leukemia (CLL) is a clonal B cell disorder of unknown origin. Accessory signals from the microenvironment are critical for the survival, expansion, and progression of malignant B cells. We found that the CLL stroma included microvascular endothelial cells (MVECs) expressing BAFF and APRIL, two TNF family members related to the T cell-associated B cell-stimulating molecule CD40L. Constitutive release of soluble BAFF and APRIL increased upon engagement of CD40 on MVECs by CD40L aberrantly expressed on CLL cells. In addition to enhancing MVEC expression of CD40, leukemic CD40L induced cleavases that elicited intracellular processing of pro-BAFF and pro-APRIL proteins in MVECs. The resulting soluble BAFF and APRIL proteins delivered survival, activation, Ig gene remodeling, and differentiation signals by stimulating CLL cells through TACI, BAFF-R, and BCMA receptors. BAFF and APRIL further amplified CLL cell survival by upregulating the expression of leukemic CD40L. Inhibition of TACI, BCMA, and BAFF-R expression on CLL cells; abrogation of CD40 expression in MVECs; or suppression of BAFF and APRIL cleavases in MVECs reduced the survival and diversification of malignant B cells. These data indicate that BAFF, APRIL, and CD40L form a CLL-enhancing bidirectional signaling network linking neoplastic B cells with the microvascular stroma.
Figures





Similar articles
-
Involvement of BAFF and APRIL in the resistance to apoptosis of B-CLL through an autocrine pathway.Blood. 2004 Jan 15;103(2):679-88. doi: 10.1182/blood-2003-02-0540. Epub 2003 Sep 22. Blood. 2004. PMID: 14504101
-
B cell activation through CD40 and IL4R ligation modulates the response of chronic lymphocytic leukaemia cells to BAFF and APRIL.Br J Haematol. 2014 Feb;164(4):570-8. doi: 10.1111/bjh.12645. Epub 2013 Nov 18. Br J Haematol. 2014. PMID: 24245956
-
Hodgkin lymphoma cells express TACI and BCMA receptors and generate survival and proliferation signals in response to BAFF and APRIL.Blood. 2007 Jan 15;109(2):729-39. doi: 10.1182/blood-2006-04-015958. Epub 2006 Sep 7. Blood. 2007. PMID: 16960154 Free PMC article.
-
Modeling the chronic lymphocytic leukemia microenvironment in vitro.Leuk Lymphoma. 2017 Feb;58(2):266-279. doi: 10.1080/10428194.2016.1204654. Epub 2016 Oct 18. Leuk Lymphoma. 2017. PMID: 27756161 Review.
-
The roles of B cell activation factor (BAFF) and a proliferation-inducing ligand (APRIL) in allergic asthma.Immunol Lett. 2020 Sep;225:25-30. doi: 10.1016/j.imlet.2020.06.001. Epub 2020 Jun 6. Immunol Lett. 2020. PMID: 32522667 Review.
Cited by
-
FCR and bevacizumab treatment in patients with relapsed chronic lymphocytic leukemia.Cancer. 2014 Nov 15;120(22):3494-501. doi: 10.1002/cncr.28910. Epub 2014 Jul 15. Cancer. 2014. PMID: 25043749 Free PMC article.
-
A novel ex vivo high-throughput assay reveals antiproliferative effects of idelalisib and ibrutinib in chronic lymphocytic leukemia.Oncotarget. 2018 May 25;9(40):26019-26031. doi: 10.18632/oncotarget.25419. eCollection 2018 May 25. Oncotarget. 2018. PMID: 29899839 Free PMC article.
-
Lymphatic endothelial cells support tumor growth in breast cancer.Sci Rep. 2014 Jul 28;4:5853. doi: 10.1038/srep05853. Sci Rep. 2014. PMID: 25068296 Free PMC article.
-
Endothelin-1 promotes survival and chemoresistance in chronic lymphocytic leukemia B cells through ETA receptor.PLoS One. 2014 Jun 5;9(6):e98818. doi: 10.1371/journal.pone.0098818. eCollection 2014. PLoS One. 2014. PMID: 24901342 Free PMC article.
-
Role of CTLA4 in the proliferation and survival of chronic lymphocytic leukemia.PLoS One. 2013 Aug 1;8(8):e70352. doi: 10.1371/journal.pone.0070352. Print 2013. PLoS One. 2013. PMID: 23936412 Free PMC article.
References
-
- Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352:804–815. - PubMed
-
- Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, Buchbinder A, Budman D, Dittmar K, Kolitz J, Lichtman SM, Schulman P, Vinciguerra VP, Rai KR, Ferrarini M, Chiorazzi N. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–1847. - PubMed
-
- Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–1854. - PubMed
-
- Wiestner A, Rosenwald A, Barry TS, Wright G, Davis RE, Henrickson SE, Zhao H, Ibbotson RE, Orchard JA, Davis Z, Stetler-Stevenson M, Raffeld M, Arthur DC, Marti GE, Wilson WH, Hamblin TJ, Oscier DG, Staudt LM. ZAP-70 expression identifies a chronic lymphocytic leukemia subtype with unmutated immunoglobulin genes, inferior clinical outcome, and distinct gene expression profile. Blood. 2003;101:4944–4951. - PubMed
-
- Furman RR, Asgary Z, Mascarenhas JO, Liou HC, Schattner EJ. Modulation of NF-kappa B activity and apoptosis in chronic lymphocytic leukemia B cells. J. Immunol. 2000;164:2200–2206. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

