High nutrient transport and cycling potential revealed in the microbial metagenome of Australian sea lion (Neophoca cinerea) faeces
- PMID: 22606263
- PMCID: PMC3350522
- DOI: 10.1371/journal.pone.0036478
High nutrient transport and cycling potential revealed in the microbial metagenome of Australian sea lion (Neophoca cinerea) faeces
Abstract
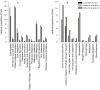
Metagenomic analysis was used to examine the taxonomic diversity and metabolic potential of an Australian sea lion (Neophoca cinerea) gut microbiome. Bacteria comprised 98% of classifiable sequences and of these matches to Firmicutes (80%) were dominant, with Proteobacteria and Actinobacteria representing 8% and 2% of matches respectively. The relative proportion of Firmicutes (80%) to Bacteriodetes (2%) is similar to that in previous studies of obese humans and obese mice, suggesting the gut microbiome may confer a predisposition towards the excess body fat that is needed for thermoregulation within the cold oceanic habitats foraged by Australian sea lions. Core metabolic functions, including carbohydrate utilisation (14%), protein metabolism (9%) and DNA metabolism (7%) dominated the metagenome, but in comparison to human and fish gut microbiomes there was a significantly higher proportion of genes involved in phosphorus metabolism (2.4%) and iron scavenging mechanisms (1%). When sea lions defecate at sea, the relatively high nutrient metabolism potential of bacteria in their faeces may accelerate the dissolution of nutrients from faecal particles, enhancing their persistence in the euphotic zone where they are available to stimulate marine production.
Conflict of interest statement
Figures




References
-
- Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-Bacterial Mutualism in the Human Intestine. Science. 2005;307:1915–1920. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

