Diversity of bifidobacteria within the infant gut microbiota
- PMID: 22606315
- PMCID: PMC3350489
- DOI: 10.1371/journal.pone.0036957
Diversity of bifidobacteria within the infant gut microbiota
Abstract
Background: The human gastrointestinal tract (GIT) represents one of the most densely populated microbial ecosystems studied to date. Although this microbial consortium has been recognized to have a crucial impact on human health, its precise composition is still subject to intense investigation. Among the GIT microbiota, bifidobacteria represent an important commensal group, being among the first microbial colonizers of the gut. However, the prevalence and diversity of members of the genus Bifidobacterium in the infant intestinal microbiota has not yet been fully characterized, while some inconsistencies exist in literature regarding the abundance of this genus.
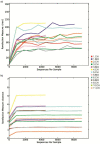
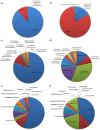
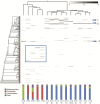
Methods/principal findings: In the current report, we assessed the complexity of the infant intestinal bifidobacterial population by analysis of pyrosequencing data of PCR amplicons derived from two hypervariable regions of the 16 S rRNA gene. Eleven faecal samples were collected from healthy infants of different geographical origins (Italy, Spain or Ireland), feeding type (breast milk or formula) and mode of delivery (vaginal or caesarean delivery), while in four cases, faecal samples of corresponding mothers were also analyzed.
Conclusions: In contrast to several previously published culture-independent studies, our analysis revealed a predominance of bifidobacteria in the infant gut as well as a profile of co-occurrence of bifidobacterial species in the infant's intestine.
Conflict of interest statement
Figures



References
-
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

