Peptide array X-linking (PAX): a new peptide-protein identification approach
- PMID: 22606326
- PMCID: PMC3351392
- DOI: 10.1371/journal.pone.0037035
Peptide array X-linking (PAX): a new peptide-protein identification approach
Abstract
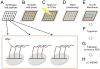
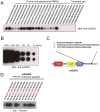
Many protein interaction domains bind short peptides based on canonical sequence consensus motifs. Here we report the development of a peptide array-based proteomics tool to identify proteins directly interacting with ligand peptides from cell lysates. Array-formatted bait peptides containing an amino acid-derived cross-linker are photo-induced to crosslink with interacting proteins from lysates of interest. Indirect associations are removed by high stringency washes under denaturing conditions. Covalently trapped proteins are subsequently identified by LC-MS/MS and screened by cluster analysis and domain scanning. We apply this methodology to peptides with different proline-containing consensus sequences and show successful identifications from brain lysates of known and novel proteins containing polyproline motif-binding domains such as EH, EVH1, SH3, WW domains. These results suggest the capacity of arrayed peptide ligands to capture and subsequently identify proteins by mass spectrometry is relatively broad and robust. Additionally, the approach is rapid and applicable to cell or tissue fractions from any source, making the approach a flexible tool for initial protein-protein interaction discovery.
Conflict of interest statement
Figures


References
-
- Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. - PubMed
-
- Yu JW, Lemmon MA. Genome-wide analysis of signaling domain function. Current opinion in chemical biology. 2003;7:103–109. - PubMed
-
- Ponting CP, Russell RR. The natural history of protein domains. Annual review of biophysics and biomolecular structure. 2002;31:45–71. - PubMed
-
- Castagnoli L, Costantini A, Dall'Armi C, Gonfloni S, Montecchi-Palazzi L, et al. Selectivity and promiscuity in the interaction network mediated by protein recognition modules. FEBS letters. 2004;567:74–79. - PubMed
-
- Takenawa T, Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol. 2007;8:37–48. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

