Gene expression profiling of liver cancer stem cells by RNA-sequencing
- PMID: 22606345
- PMCID: PMC3351419
- DOI: 10.1371/journal.pone.0037159
Gene expression profiling of liver cancer stem cells by RNA-sequencing
Abstract
Background: Accumulating evidence supports that tumor growth and cancer relapse are driven by cancer stem cells. Our previous work has demonstrated the existence of CD90(+) liver cancer stem cells (CSCs) in hepatocellular carcinoma (HCC). Nevertheless, the characteristics of these cells are still poorly understood. In this study, we employed a more sensitive RNA-sequencing (RNA-Seq) to compare the gene expression profiling of CD90(+) cells sorted from tumor (CD90(+)CSCs) with parallel non-tumorous liver tissues (CD90(+)NTSCs) and elucidate the roles of putative target genes in hepatocarcinogenesis.
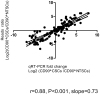
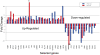
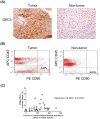
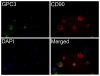
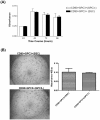
Methodology/principal findings: CD90(+) cells were sorted respectively from tumor and adjacent non-tumorous human liver tissues using fluorescence-activated cell sorting. The amplified RNAs of CD90(+) cells from 3 HCC patients were subjected to RNA-Seq analysis. A differential gene expression profile was established between CD90(+)CSCs and CD90(+)NTSCs, and validated by quantitative real-time PCR (qRT-PCR) on the same set of amplified RNAs, and further confirmed in an independent cohort of 12 HCC patients. Five hundred genes were differentially expressed (119 up-regulated and 381 down-regulated genes) between CD90(+)CSCs and CD90(+)NTSCs. Gene ontology analysis indicated that the over-expressed genes in CD90(+)CSCs were associated with inflammation, drug resistance and lipid metabolism. Among the differentially expressed genes, glypican-3 (GPC3), a member of glypican family, was markedly elevated in CD90(+)CSCs compared to CD90(+)NTSCs. Immunohistochemistry demonstrated that GPC3 was highly expressed in forty-two human liver tumor tissues but absent in adjacent non-tumorous liver tissues. Flow cytometry indicated that GPC3 was highly expressed in liver CD90(+)CSCs and mature cancer cells in liver cancer cell lines and human liver tumor tissues. Furthermore, GPC3 expression was positively correlated with the number of CD90(+)CSCs in liver tumor tissues.
Conclusions/significance: The identified genes, such as GPC3 that are distinctly expressed in liver CD90(+)CSCs, may be promising gene candidates for HCC therapy without inducing damages to normal liver stem cells.
Conflict of interest statement
Figures
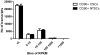



References
-
- Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346. - PubMed
-
- Lo CM, Ngan H, Tso WK, Liu CL, Lam CM, et al. Randomized controlled trial of transarterial lipiodol chemoembolization for unresectable hepatocellular carcinoma. Hepatology. 2002;35:1164–1171. - PubMed
-
- Ma S, Jiao B, Liu X, Yi H, Kong D, et al. Cancer Treat Rev; 2009. Approach to radiation therapy in hepatocellular carcinoma. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

