MicroRNA regulation of human protease genes essential for influenza virus replication
- PMID: 22606348
- PMCID: PMC3351457
- DOI: 10.1371/journal.pone.0037169
MicroRNA regulation of human protease genes essential for influenza virus replication
Abstract
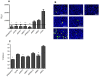
Influenza A virus causes seasonal epidemics and periodic pandemics threatening the health of millions of people each year. Vaccination is an effective strategy for reducing morbidity and mortality, and in the absence of drug resistance, the efficacy of chemoprophylaxis is comparable to that of vaccines. However, the rapid emergence of drug resistance has emphasized the need for new drug targets. Knowledge of the host cell components required for influenza replication has been an area targeted for disease intervention. In this study, the human protease genes required for influenza virus replication were determined and validated using RNA interference approaches. The genes validated as critical for influenza virus replication were ADAMTS7, CPE, DPP3, MST1, and PRSS12, and pathway analysis showed these genes were in global host cell pathways governing inflammation (NF-κB), cAMP/calcium signaling (CRE/CREB), and apoptosis. Analyses of host microRNAs predicted to govern expression of these genes showed that eight miRNAs regulated gene expression during virus replication. These findings identify unique host genes and microRNAs important for influenza replication providing potential new targets for disease intervention strategies.
Conflict of interest statement
Figures
References
-
- Kandel R, Hartshorn KL. Prophylaxis and treatment of influenza virus infection. BioDrugs. 2001;15:303–323. - PubMed
-
- Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289:179–186. - PubMed
-
- Hale BG, Randall RE, Ortin J, Jackson D. The multifunctional NS1 protein of influenza A viruses. J Gen Virol. 2008;89:2359–2376. - PubMed
-
- Basler CF. Influenza viruses: basic biology and potential drug targets. Infect Disord Drug Targets. 2007;7:282–293. - PubMed
-
- Betakova T. M2 protein-a proton channel of influenza A virus. Curr Pharm Des. 2007;13:3231–3235. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

