Genetic variation and metabolic pathway intricacy govern the active compound content and quality of the Chinese medicinal plant Lonicera japonica thunb
- PMID: 22607188
- PMCID: PMC3443457
- DOI: 10.1186/1471-2164-13-195
Genetic variation and metabolic pathway intricacy govern the active compound content and quality of the Chinese medicinal plant Lonicera japonica thunb
Abstract
Background: Traditional Chinese medicine uses various herbs for the treatment of various diseases for thousands of years and it is now time to assess the characteristics and effectiveness of these medicinal plants based on modern genetic and molecular tools. The herb Flos Lonicerae Japonicae (FLJ or Lonicera japonica Thunb.) is used as an anti-inflammatory agent but the chemical quality of FLJ and its medicinal efficacy has not been consistent. Here, we analyzed the transcriptomes and metabolic pathways to evaluate the active medicinal compounds in FLJ and hope that this approach can be used for a variety of medicinal herbs in the future.
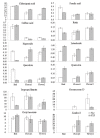
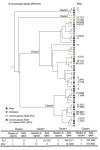
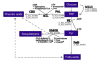
Results: We assess transcriptomic differences between FLJ and L. japonica Thunb. var. chinensis (Watts) (rFLJ), which may explain the variable medicinal effects. We acquired transcriptomic data (over 100 million reads) from the two herbs, using RNA-seq method and the Illumina GAII platform. The transcriptomic profiles contain over 6,000 expressed sequence tags (ESTs) for each of the three flower development stages from FLJ, as well as comparable amount of ESTs from the rFLJ flower bud. To elucidate enzymatic divergence on biosynthetic pathways between the two varieties, we correlated genes and their expression profiles to known metabolic activities involving the relevant active compounds, including phenolic acids, flavonoids, terpenoids, and fatty acids. We also analyzed the diversification of genes that process the active compounds to distinguish orthologs and paralogs together with the pathways concerning biosynthesis of phenolic acid and its connections with other related pathways.
Conclusions: Our study provides both an initial description of gene expression profiles in flowers of FLJ and its counterfeit rFLJ and the enzyme pool that can be used to evaluate FLJ quality. Detailed molecular-level analyses allow us to decipher the relationship between metabolic pathways involved in processing active medicinal compounds and gene expressions of their processing enzymes. Our evolutionary analysis revealed specific functional divergence of orthologs and paralogs, which lead to variation in gene functions that govern the profile of active compounds.
Figures





References
-
- Hong SZJ, Yang G, Luo X. Earliest report of Flos Lonicerae Japonicae and its medical organ. Zhongyaocai. 1997;20:2.
-
- Xiang T, Xiong QB, Ketut AI, Tezuka Y, Nagaoka T, Wu LJ, Kadota S. Studies on the hepatocyte protective activity and the structure-activity relationships of quinic acid and caffeic acid derivatives from the flower buds of Lonicera bournei. Planta Med. 2001;67:322–325. doi: 10.1055/s-2001-14337. - DOI - PubMed
-
- Yoo HJ, Kang HJ, Song YS, Park EH, Lim CJ. Anti-angiogenic, antinociceptive and anti-inflammatory activities of Lonicera japonica extract. J Pharm Pharmacol. 2008;60:779–786. - PubMed
-
- Li P, Qi LW, Chen CY. Structural characterization and identification of iridoid glycosides, saponins, phenolic acids and flavonoids in Flos Lonicerae Japonicae by a fast liquid chromatography method with diode-array detection and time-of-flight mass spectrometry. Rapid Commun Mass Sp. 2009;23:3227–3242. doi: 10.1002/rcm.4245. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

