From networks of protein interactions to networks of functional dependencies
- PMID: 22607727
- PMCID: PMC3434018
- DOI: 10.1186/1752-0509-6-44
From networks of protein interactions to networks of functional dependencies
Abstract
Background: As protein-protein interactions connect proteins that participate in either the same or different functions, networks of interacting and functionally annotated proteins can be converted into process graphs of inter-dependent function nodes (each node corresponding to interacting proteins with the same functional annotation). However, as proteins have multiple annotations, the process graph is non-redundant, if only proteins participating directly in a given function are included in the related function node.
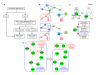
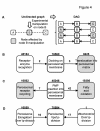
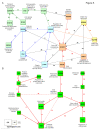
Results: Reasoning that topological features (e.g., clusters of highly inter-connected proteins) might help approaching structured and non-redundant understanding of molecular function, an algorithm was developed that prioritizes inclusion of proteins into the function nodes that best overlap protein clusters. Specifically, the algorithm identifies function nodes (and their mutual relations), based on the topological analysis of a protein interaction network, which can be related to various biological domains, such as cellular components (e.g., peroxisome and cellular bud) or biological processes (e.g., cell budding) of the model organism S. cerevisiae.
Conclusions: The method we have described allows converting a protein interaction network into a non-redundant process graph of inter-dependent function nodes. The examples we have described show that the resulting graph allows researchers to formulate testable hypotheses about dependencies among functions and the underlying mechanisms.
Figures


Similar articles
-
L-GRAAL: Lagrangian graphlet-based network aligner.Bioinformatics. 2015 Jul 1;31(13):2182-9. doi: 10.1093/bioinformatics/btv130. Epub 2015 Feb 28. Bioinformatics. 2015. PMID: 25725498 Free PMC article.
-
Evaluation of clustering algorithms for protein-protein interaction networks.BMC Bioinformatics. 2006 Nov 6;7:488. doi: 10.1186/1471-2105-7-488. BMC Bioinformatics. 2006. PMID: 17087821 Free PMC article.
-
Visualization and analysis of the complexome network of Saccharomyces cerevisiae.J Proteome Res. 2011 Oct 7;10(10):4744-56. doi: 10.1021/pr200548c. Epub 2011 Sep 7. J Proteome Res. 2011. PMID: 21842913
-
Sharing the wealth: peroxisome inheritance in budding yeast.Biochim Biophys Acta. 2006 Dec;1763(12):1669-77. doi: 10.1016/j.bbamcr.2006.08.015. Epub 2006 Aug 22. Biochim Biophys Acta. 2006. PMID: 17005268 Review.
-
Systematic curation of protein and genetic interaction data for computable biology.BMC Biol. 2013 Apr 15;11:43. doi: 10.1186/1741-7007-11-43. BMC Biol. 2013. PMID: 23587305 Free PMC article. Review. No abstract available.
Cited by
-
Interlog protein network: an evolutionary benchmark of protein interaction networks for the evaluation of clustering algorithms.BMC Bioinformatics. 2015 Oct 5;16:319. doi: 10.1186/s12859-015-0755-1. BMC Bioinformatics. 2015. PMID: 26437714 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

