Molecular modeling of inhibitors of human DNA methyltransferase with a crystal structure: discovery of a novel DNMT1 inhibitor
- PMID: 22607757
- PMCID: PMC3837394
- DOI: 10.1016/B978-0-12-398312-1.00008-1
Molecular modeling of inhibitors of human DNA methyltransferase with a crystal structure: discovery of a novel DNMT1 inhibitor
Abstract
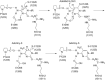
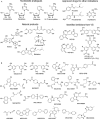
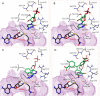
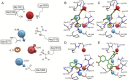
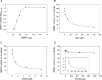
DNA methyltransferases (DNMTs) are promising epigenetic targets for the development of novel anticancer drugs and other diseases. Molecular modeling and experimental approaches are being used to identify and develop inhibitors of human DNMTs. Most of the computational efforts conducted so far with DNMT1 employ homology models of the enzyme. Recently, a crystallographic structure of the methyltransferase domain of human DNMT1 bound to unmethylated DNA was published. Following on our previous computational and experimental studies with DNMTs, we herein present molecular dynamics of the crystal structure of human DNMT1. Docking studies of established DNMT1 inhibitors with the crystal structure gave rise to a structure-based pharmacophore model that suggests key interactions of the inhibitors with the catalytic binding site. Results had a good agreement with the docking and pharmacophore models previously developed using a homology model of the catalytic domain of DNMT1. The docking protocol was able to distinguish active DNMT1 inhibitors from, for example, experimentally known inactive DNMT1 inhibitors. As part of our efforts to identify novel inhibitors of DNMT1, we conducted the experimental characterization of aurintricarboxylic acid (ATA) that in preliminary docking studies showed promising activity. ATA had a submicromolar inhibition (IC(50)=0.68 μM) against DNMT1. ATA was also evaluated for Dnmt3a inhibition showing an IC(50)=1.4 μM. This chapter illustrates the synergy from integrating molecular modeling and experimental methods to further advance the discovery of novel candidates for epigenetic therapies.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures


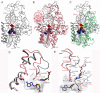

References
-
- Caulfield T., Medina-Franco J.L. Molecular dynamics simulations of human DNA methyltransferase 3B with selective inhibitor nanaomycin A. J. Struct. Biol. 2011;176:185–191. - PubMed
-
- Chen T.P., Hevi S., Gay F., Tsujimoto N., He T., Zhang B.L. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat. Genet. 2007;39:391–396. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

