A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach
- PMID: 22607806
- PMCID: PMC3358704
- DOI: 10.1016/j.chom.2012.04.007
A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach
Abstract
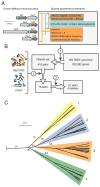
Sophisticated mechanisms are employed to facilitate information exchange between interfacing bacteria. A type VI secretion system (T6SS) of Pseudomonas aeruginosa was shown to deliver cell wall-targeting effectors to neighboring cells. However, the generality of bacteriolytic effectors and, moreover, of antibacterial T6S remained unknown. Using parameters derived from experimentally validated bacterial T6SS effectors we identified a phylogenetically disperse superfamily of T6SS-associated peptidoglycan-degrading effectors. The effectors separate into four families composed of peptidoglycan amidase enzymes of differing specificities. Effectors strictly co-occur with cognate immunity proteins, indicating that self-intoxication is a general property of antibacterial T6SSs and effector delivery by the system exerts a strong selective pressure in nature. The presence of antibacterial effectors in a plethora of organisms, including many that inhabit or infect polymicrobial niches in the human body, suggests that the system could mediate interbacterial interactions of both environmental and clinical significance.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures




References
-
- Bateman A, Rawlings ND. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends in biochemical sciences. 2003;28:234–237. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

