Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions
- PMID: 22607975
- PMCID: PMC3361558
- DOI: 10.1016/j.molcel.2012.03.030
Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions
Abstract
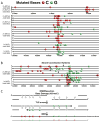
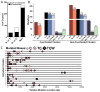
Mutations are typically perceived as random, independent events. We describe here nonrandom clustered mutations in yeast and in human cancers. Genome sequencing of yeast grown under chronic alkylation damage identified mutation clusters that extend up to 200 kb. A predominance of "strand-coordinated" changes of either cytosines or guanines in the same strand, mutation patterns, and genetic controls indicated that simultaneous mutations were generated by base alkylation in abnormally long single-strand DNA (ssDNA) formed at double-strand breaks (DSBs) and replication forks. Significantly, we found mutation clusters with analogous features in sequenced human cancers. Strand-coordinated clusters of mutated cytosines or guanines often resided near chromosome rearrangement breakpoints and were highly enriched with a motif targeted by APOBEC family cytosine-deaminases, which strongly prefer ssDNA. These data indicate that hypermutation via multiple simultaneous changes in randomly formed ssDNA is a general phenomenon that may be an important mechanism producing rapid genetic variation.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures



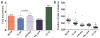
Comment in
-
Tumor archaeology reveals that mutations love company.Cell. 2012 May 25;149(5):959-61. doi: 10.1016/j.cell.2012.05.010. Cell. 2012. PMID: 22632962 No abstract available.
-
Genomic instability. Mutagenic clusters.Nat Rev Cancer. 2012 Jun 22;12(7):452-3. doi: 10.1038/nrc3306. Nat Rev Cancer. 2012. PMID: 22722398
References
-
- Beale RC, Petersen-Mahrt SK, Watt IN, Harris RS, Rada C, Neuberger MS. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J Mol Biol. 2004;337:585–596. - PubMed
-
- Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat Res. 1990;231:11–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

