Molecular diagnostics for congenital hearing loss including 15 deafness genes using a next generation sequencing platform
- PMID: 22607986
- PMCID: PMC3443074
- DOI: 10.1186/1755-8794-5-17
Molecular diagnostics for congenital hearing loss including 15 deafness genes using a next generation sequencing platform
Abstract
Background: Hereditary hearing loss (HL) can originate from mutations in one of many genes involved in the complex process of hearing. Identification of the genetic defects in patients is currently labor intensive and expensive. While screening with Sanger sequencing for GJB2 mutations is common, this is not the case for the other known deafness genes (> 60). Next generation sequencing technology (NGS) has the potential to be much more cost efficient. Published methods mainly use hybridization based target enrichment procedures that are time saving and efficient, but lead to loss in sensitivity. In this study we used a semi-automated PCR amplification and NGS in order to combine high sensitivity, speed and cost efficiency.
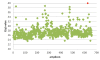
Results: In this proof of concept study, we screened 15 autosomal recessive deafness genes in 5 patients with congenital genetic deafness. 646 specific primer pairs for all exons and most of the UTR of the 15 selected genes were designed using primerXL. Using patient specific identifiers, all amplicons were pooled and analyzed using the Roche 454 NGS technology. Three of these patients are members of families in which a region of interest has previously been characterized by linkage studies. In these, we were able to identify two new mutations in CDH23 and OTOF. For another patient, the etiology of deafness was unclear, and no causal mutation was found. In a fifth patient, included as a positive control, we could confirm a known mutation in TMC1.
Conclusions: We have developed an assay that holds great promise as a tool for screening patients with familial autosomal recessive nonsyndromal hearing loss (ARNSHL). For the first time, an efficient, reliable and cost effective genetic test, based on PCR enrichment, for newborns with undiagnosed deafness is available.
Figures

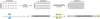
References
-
- The Hereditary Hearing Loss Homepage. [ http://hereditaryhearingloss.org]
-
- Shearer AE, Deluca AP, Hildebrand MS, Taylor KR, Gurrola J, Scherer S. et al.Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21104–21109. doi: 10.1073/pnas.1012989107. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

