The life history of 21 breast cancers
- PMID: 22608083
- PMCID: PMC3428864
- DOI: 10.1016/j.cell.2012.04.023
The life history of 21 breast cancers
Erratum in
- Cell. 2015 Aug 13;162(4):924
Abstract
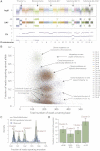
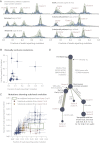
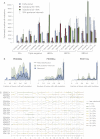
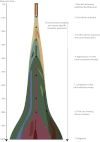
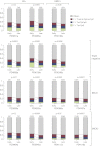
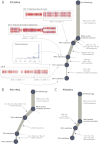
Cancer evolves dynamically as clonal expansions supersede one another driven by shifting selective pressures, mutational processes, and disrupted cancer genes. These processes mark the genome, such that a cancer's life history is encrypted in the somatic mutations present. We developed algorithms to decipher this narrative and applied them to 21 breast cancers. Mutational processes evolve across a cancer's lifespan, with many emerging late but contributing extensive genetic variation. Subclonal diversification is prominent, and most mutations are found in just a fraction of tumor cells. Every tumor has a dominant subclonal lineage, representing more than 50% of tumor cells. Minimal expansion of these subclones occurs until many hundreds to thousands of mutations have accumulated, implying the existence of long-lived, quiescent cell lineages capable of substantial proliferation upon acquisition of enabling genomic changes. Expansion of the dominant subclone to an appreciable mass may therefore represent the final rate-limiting step in a breast cancer's development, triggering diagnosis.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures









Comment in
-
Tumor archaeology reveals that mutations love company.Cell. 2012 May 25;149(5):959-61. doi: 10.1016/j.cell.2012.05.010. Cell. 2012. PMID: 22632962 No abstract available.
References
-
- Anderson K., Lutz C., van Delft F.W., Bateman C.M., Guo Y., Colman S.M., Kempski H., Moorman A.V., Titley I., Swansbury J. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. - PubMed
Supplemental References
-
- Baumbusch, L.O., Aarøe, J., Johansen, F.E., Hicks, J., Sun, H., Bruhn, L., Gunderson, K., Naume, B., Kristensen, V.N., Liestøl, K., et al. (2008). Comparison of the Agilent, ROMA/NimbleGen and Illumina platforms for classification of copy number alterations in human breast tumors. BMC Genomics 9, 379. - PMC - PubMed
-
- Dunson, D.B. (2010). Nonparametric Bayes applications to biostatistics. In Bayesian Nonparametrics, N.L. Hjort, C. Holmes, P. Müller, and S.G. Walker, eds. (Cambridge: Cambridge University Press).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

