Mutational processes molding the genomes of 21 breast cancers
- PMID: 22608084
- PMCID: PMC3414841
- DOI: 10.1016/j.cell.2012.04.024
Mutational processes molding the genomes of 21 breast cancers
Abstract
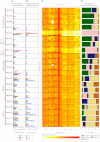
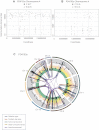
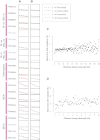
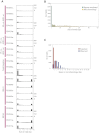
All cancers carry somatic mutations. The patterns of mutation in cancer genomes reflect the DNA damage and repair processes to which cancer cells and their precursors have been exposed. To explore these mechanisms further, we generated catalogs of somatic mutation from 21 breast cancers and applied mathematical methods to extract mutational signatures of the underlying processes. Multiple distinct single- and double-nucleotide substitution signatures were discernible. Cancers with BRCA1 or BRCA2 mutations exhibited a characteristic combination of substitution mutation signatures and a distinctive profile of deletions. Complex relationships between somatic mutation prevalence and transcription were detected. A remarkable phenomenon of localized hypermutation, termed "kataegis," was observed. Regions of kataegis differed between cancers but usually colocalized with somatic rearrangements. Base substitutions in these regions were almost exclusively of cytosine at TpC dinucleotides. The mechanisms underlying most of these mutational signatures are unknown. However, a role for the APOBEC family of cytidine deaminases is proposed.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures




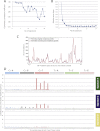

Comment in
-
Tumor archaeology reveals that mutations love company.Cell. 2012 May 25;149(5):959-61. doi: 10.1016/j.cell.2012.05.010. Cell. 2012. PMID: 22632962 No abstract available.
References
-
- Berry M.W., Browne M., Langville A.N., Pauca V.P., Plemmons R.J. Algorithms and applications for approximate nonnegative matrix factorization. Comput. Stat. Data Anal. 2007;52:155–173.
Supplemental References
-
- Campbell, P.J., Stephens, P.J., Pleasance, E.D., O'Meara, S., Li, H., Santarius, T., Stebbings, L.A., Leroy, C., Edkins, S., Hardy, C., et al. (2008). Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet. 40, 722–729. - PMC - PubMed
-
- Lee, D.D., and Seung, H.S. (1999). Learning the parts of objects by non-negative matrix factorization. Nature 401, 788–791. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

