Comparative genomic analyses of 17 clinical isolates of Gardnerella vaginalis provide evidence of multiple genetically isolated clades consistent with subspeciation into genovars
- PMID: 22609915
- PMCID: PMC3416530
- DOI: 10.1128/JB.00056-12
Comparative genomic analyses of 17 clinical isolates of Gardnerella vaginalis provide evidence of multiple genetically isolated clades consistent with subspeciation into genovars
Abstract
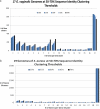
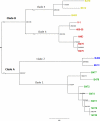
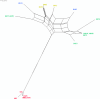
Gardnerella vaginalis is associated with a spectrum of clinical conditions, suggesting high degrees of genetic heterogeneity among stains. Seventeen G. vaginalis isolates were subjected to a battery of comparative genomic analyses to determine their level of relatedness. For each measure, the degree of difference among the G. vaginalis strains was the highest observed among 23 pathogenic bacterial species for which at least eight genomes are available. Genome sizes ranged from 1.491 to 1.716 Mb; GC contents ranged from 41.18% to 43.40%; and the core genome, consisting of only 746 genes, makes up only 51.6% of each strain's genome on average and accounts for only 27% of the species supragenome. Neighbor-grouping analyses, using both distributed gene possession data and core gene allelic data, each identified two major sets of strains, each of which is composed of two groups. Each of the four groups has its own characteristic genome size, GC ratio, and greatly expanded core gene content, making the genomic diversity of each group within the range for other bacterial species. To test whether these 4 groups corresponded to genetically isolated clades, we inferred the phylogeny of each distributed gene that was present in at least two strains and absent in at least two strains; this analysis identified frequent homologous recombination within groups but not between groups or sets. G. vaginalis appears to include four nonrecombining groups/clades of organisms with distinct gene pools and genomic properties, which may confer distinct ecological properties. Consequently, it may be appropriate to treat these four groups as separate species.
Figures


References
-
- Amsel R, et al. 1983. Nonspecific vaginitis: diagnostic criteria and microbial and epidemiologic associations. Am. J. Med. 74:14–22 - PubMed
-
- Anukam KC, Osazuwa EO, Ahonkhai I, Reid GG. 2006. Lactobacillus vaginal microbiota of women attending a reproductive health care service in Benin City, Nigeria. Sex. Transm. Dis. 33:59–62 - PubMed
-
- Aroutcheva AA, Simoes JA, Behbakht K, Faro S. 2001. Gardnerella vaginalis isolate from patients with bacterial vaginosis and from patients with healthy vaginal ecosystems. Clin. Infect. Dis. 33:1022–1027 - PubMed
-
- Ashida H, et al. 2009. Two distinct alpha-L-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology 19:1010–1017 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

