Single-CpG-resolution methylome analysis identifies clinicopathologically aggressive CpG island methylator phenotype clear cell renal cell carcinomas
- PMID: 22610075
- PMCID: PMC3418891
- DOI: 10.1093/carcin/bgs177
Single-CpG-resolution methylome analysis identifies clinicopathologically aggressive CpG island methylator phenotype clear cell renal cell carcinomas
Abstract
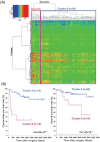
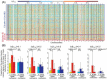
To clarify the significance of DNA methylation alterations during renal carcinogenesis, methylome analysis using single-CpG-resolution Infinium array was performed on 29 normal renal cortex tissue (C) samples, 107 non-cancerous renal cortex tissue (N) samples obtained from patients with clear cell renal cell carcinomas (RCCs) and 109 tumorous tissue (T) samples. DNA methylation levels at 4830 CpG sites were already altered in N samples compared with C samples. Unsupervised hierarchical clustering analysis based on DNA methylation levels at the 801 CpG sites, where DNA methylation alterations had occurred in N samples and were inherited by and strengthened in T samples, clustered clear cell RCCs into Cluster A (n = 90) and Cluster B (n = 14). Clinicopathologically aggressive tumors were accumulated in Cluster B, and the cancer-free and overall survival rates of patients in this cluster were significantly lower than those of patients in Cluster A. Clear cell RCCs in Cluster B were characterized by accumulation of DNA hypermethylation on CpG islands and considered to be CpG island methylator phenotype (CIMP)-positive cancers. DNA hypermethylation of the CpG sites on the FAM150A, GRM6, ZNF540, ZFP42, ZNF154, RIMS4, PCDHAC1, KHDRBS2, ASCL2, KCNQ1, PRAC, WNT3A, TRH, FAM78A, ZNF671, SLC13A5 and NKX6-2 genes became hallmarks of CIMP in RCCs. On the other hand, Cluster A was characterized by genome-wide DNA hypomethylation. These data indicated that DNA methylation alterations at precancerous stages may determine tumor aggressiveness and patient outcome. Accumulation of DNA hypermethylation on CpG islands and genome-wide DNA hypomethylation may each underlie distinct pathways of renal carcinogenesis.
Figures
References
-
- Ljungberg B., et al. (2011). The epidemiology of renal cell carcinoma Eur. Urol. 60 615–621 - PubMed
-
- Baldewijns M.M., et al. (2010). VHL and HIF signalling in renal cell carcinogenesis J. Pathol., 221 125–138 - PubMed
-
- Barrett I.P. (2010). Cancer genome analysis informatics Methods Mol. Biol. 628 75–102 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

