Coiled coil domain-containing protein 56 (CCDC56) is a novel mitochondrial protein essential for cytochrome c oxidase function
- PMID: 22610097
- PMCID: PMC3397844
- DOI: 10.1074/jbc.M112.343764
Coiled coil domain-containing protein 56 (CCDC56) is a novel mitochondrial protein essential for cytochrome c oxidase function
Abstract
In Drosophila melanogaster, the mitochondrial transcription factor B1 (d-mtTFB1) transcript contains in its 5'-untranslated region a conserved upstream open reading frame denoted as CG42630 in FlyBase. We demonstrate that CG42630 encodes a novel protein, the coiled coil domain-containing protein 56 (CCDC56), conserved in metazoans. We show that Drosophila CCDC56 protein localizes to mitochondria and contains 87 amino acids in flies and 106 in humans with the two proteins sharing 42% amino acid identity. We show by rapid amplification of cDNA ends and Northern blotting that Drosophila CCDC56 protein and mtTFB1 are encoded on a bona fide bicistronic transcript. We report the generation and characterization of two ccdc56 knock-out lines in Drosophila carrying the ccdc56(D6) and ccdc56(D11) alleles. Lack of the CCDC56 protein in flies induces a developmental delay and 100% lethality by arrest of larval development at the third instar. ccdc56 knock-out larvae show a significant decrease in the level of fully assembled cytochrome c oxidase (COX) and in its activity, suggesting a defect in complex assembly; the activity of the other oxidative phosphorylation complexes remained either unaffected or increased in the ccdc56 knock-out larvae. The lethal phenotype and the decrease in COX were partially rescued by reintroduction of a wild-type UAS-ccdc56 transgene. These results indicate an important role for CCDC56 in the oxidative phosphorylation system and in particular in COX function required for proper development in D. melanogaster. We propose CCDC56 as a candidate factor required for COX biogenesis/assembly.
Figures

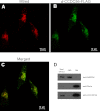

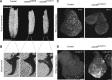



References
-
- Tsukihara T., Aoyama H., Yamashita E., Tomizaki T., Yamaguchi H., Shinzawa-Itoh K., Nakashima R., Yaono R., Yoshikawa S. (1995) Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 Å. Science 269, 1069–1074 - PubMed
-
- Galati D., Srinivasan S., Raza H., Prabu S. K., Hardy M., Chandran K., Lopez M., Kalyanaraman B., Avadhani N. G. (2009) Role of nuclear-encoded subunit Vb in the assembly and stability of cytochrome c oxidase complex: implications in mitochondrial dysfunction and ROS production. Biochem. J. 420, 439–449 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

