Inferring direct DNA binding from ChIP-seq
- PMID: 22610855
- PMCID: PMC3458523
- DOI: 10.1093/nar/gks433
Inferring direct DNA binding from ChIP-seq
Abstract
Genome-wide binding data from transcription factor ChIP-seq experiments is the best source of information for inferring the relative DNA-binding affinity of these proteins in vivo. However, standard motif enrichment analysis and motif discovery approaches sometimes fail to correctly identify the binding motif for the ChIP-ed factor. To overcome this problem, we propose 'central motif enrichment analysis' (CMEA), which is based on the observation that the positional distribution of binding sites matching the direct-binding motif tends to be unimodal, well centered and maximal in the precise center of the ChIP-seq peak regions. We describe a novel visualization and statistical analysis tool--CentriMo--that identifies the region of maximum central enrichment in a set of ChIP-seq peak regions and displays the positional distributions of predicted sites. Using CentriMo for motif enrichment analysis, we provide evidence that one transcription factor (Nanog) has different binding affinity in vivo than in vitro, that another binds DNA cooperatively (E2f1), and confirm the in vivo affinity of NFIC, rescuing a difficult ChIP-seq data set. In another data set, CentriMo strongly suggests that there is no evidence of direct DNA binding by the ChIP-ed factor (Smad1). CentriMo is now part of the MEME Suite software package available at http://meme.nbcr.net. All data and output files presented here are available at: http://research.imb.uq.edu.au/t.bailey/sd/Bailey2011a.
Figures


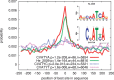
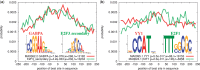


References
-
- Stormo GD. Information content and free energy in DNA–protein interactions. J. Theor. Biol. 1998;195:135–137. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

