Evolutionary history of LTR retrotransposon chromodomains in plants
- PMID: 22611377
- PMCID: PMC3350952
- DOI: 10.1155/2012/874743
Evolutionary history of LTR retrotransposon chromodomains in plants
Abstract
Chromodomain-containing LTR retrotransposons are one of the most successful groups of mobile elements in plant genomes. Previously, we demonstrated that two types of chromodomains (CHDs) are carried by plant LTR retrotransposons. Chromodomains from group I (CHD_I) were detected only in Tcn1-like LTR retrotransposons from nonseed plants such as mosses (including the model moss species Physcomitrella) and lycophytes (the Selaginella species). LTR retrotransposon chromodomains from group II (CHD_II) have been described from a wide range of higher plants. In the present study, we performed computer-based mining of plant LTR retrotransposon CHDs from diverse plants with an emphasis on spike-moss Selaginella. Our extended comparative and phylogenetic analysis demonstrated that two types of CHDs are present only in the Selaginella genome, which puts this species in a unique position among plants. It appears that a transition from CHD_I to CHD_II and further diversification occurred in the evolutionary history of plant LTR retrotransposons at approximately 400 MYA and most probably was associated with the evolution of chromatin organization.
Figures
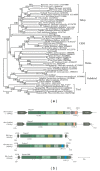
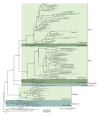
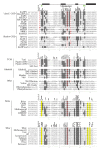
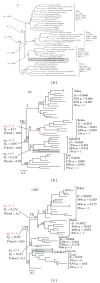
Similar articles
-
Evolutionary genomics revealed interkingdom distribution of Tcn1-like chromodomain-containing Gypsy LTR retrotransposons among fungi and plants.BMC Genomics. 2010 Apr 8;11:231. doi: 10.1186/1471-2164-11-231. BMC Genomics. 2010. PMID: 20377908 Free PMC article.
-
Chromodomains and LTR retrotransposons in plants.Commun Integr Biol. 2009;2(2):158-62. doi: 10.4161/cib.7702. Commun Integr Biol. 2009. PMID: 19513271 Free PMC article.
-
Novel clades of chromodomain-containing Gypsy LTR retrotransposons from mosses (Bryophyta).Plant J. 2008 Nov;56(4):562-74. doi: 10.1111/j.1365-313X.2008.03621.x. Epub 2008 Aug 11. Plant J. 2008. PMID: 18643967
-
LTR retrotransposons and flowering plant genome size: emergence of the increase/decrease model.Cytogenet Genome Res. 2005;110(1-4):91-107. doi: 10.1159/000084941. Cytogenet Genome Res. 2005. PMID: 16093661 Review.
-
LTR-retrotransposons in plants: Engines of evolution.Gene. 2017 Aug 30;626:14-25. doi: 10.1016/j.gene.2017.04.051. Epub 2017 May 2. Gene. 2017. PMID: 28476688 Review.
Cited by
-
Convergent evolution of tRNA gene targeting preferences in compact genomes.Mob DNA. 2016 Aug 31;7(1):17. doi: 10.1186/s13100-016-0073-9. eCollection 2016. Mob DNA. 2016. PMID: 27583033 Free PMC article.
-
The discovery of a key prenyltransferase gene assisted by a chromosome-level Epimedium pubescens genome.Front Plant Sci. 2022 Nov 14;13:1034943. doi: 10.3389/fpls.2022.1034943. eCollection 2022. Front Plant Sci. 2022. PMID: 36452098 Free PMC article.
-
Retrotransposons in Plant Genomes: Structure, Identification, and Classification through Bioinformatics and Machine Learning.Int J Mol Sci. 2019 Aug 6;20(15):3837. doi: 10.3390/ijms20153837. Int J Mol Sci. 2019. PMID: 31390781 Free PMC article. Review.
-
sRNAs as possible regulators of retrotransposon activity in Cryptococcus gattii VGII.BMC Genomics. 2017 Apr 12;18(1):294. doi: 10.1186/s12864-017-3688-4. BMC Genomics. 2017. PMID: 28403818 Free PMC article.
-
Convergence of retrotransposons in oomycetes and plants.Mob DNA. 2017 Mar 14;8:4. doi: 10.1186/s13100-017-0087-y. eCollection 2017. Mob DNA. 2017. PMID: 28293305 Free PMC article.
References
-
- Brehm A, Tufteland KR, Aasland R, Becker PB. The many colours of chromodomains. BioEssays. 2004;26(2):133–140. - PubMed
LinkOut - more resources
Full Text Sources

