Elastic network normal modes provide a basis for protein structure refinement
- PMID: 22612113
- PMCID: PMC3365912
- DOI: 10.1063/1.4710986
Elastic network normal modes provide a basis for protein structure refinement
Abstract
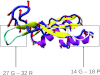
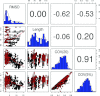
It is well recognized that thermal motions of atoms in the protein native state, the fluctuations about the minimum of the global free energy, are well reproduced by the simple elastic network models (ENMs) such as the anisotropic network model (ANM). Elastic network models represent protein dynamics as vibrations of a network of nodes (usually represented by positions of the heavy atoms or by the C(α) atoms only for coarse-grained representations) in which the spatially close nodes are connected by harmonic springs. These models provide a reliable representation of the fluctuational dynamics of proteins and RNA, and explain various conformational changes in protein structures including those important for ligand binding. In the present paper, we study the problem of protein structure refinement by analyzing thermal motions of proteins in non-native states. We represent the conformational space close to the native state by a set of decoys generated by the I-TASSER protein structure prediction server utilizing template-free modeling. The protein substates are selected by hierarchical structure clustering. The main finding is that thermal motions for some substates, overlap significantly with the deformations necessary to reach the native state. Additionally, more mobile residues yield higher overlaps with the required deformations than do the less mobile ones. These findings suggest that structural refinement of poorly resolved protein models can be significantly enhanced by reduction of the conformational space to the motions imposed by the dominant normal modes.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

