Frequency and genetic characterization of V(DD)J recombinants in the human peripheral blood antibody repertoire
- PMID: 22612413
- PMCID: PMC3449247
- DOI: 10.1111/j.1365-2567.2012.03605.x
Frequency and genetic characterization of V(DD)J recombinants in the human peripheral blood antibody repertoire
Abstract
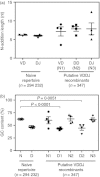
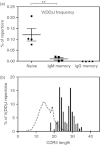
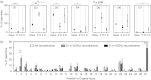
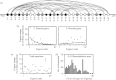
Antibody heavy-chain recombination that results in the incorporation of multiple diversity (D) genes, although uncommon, contributes substantially to the diversity of the human antibody repertoire. Such recombination allows the generation of heavy chain complementarity determining region 3 (HCDR3) regions of extreme length and enables junctional regions that, because of the nucleotide bias of N-addition regions, are difficult to produce through normal V(D)J recombination. Although this non-classical recombination process has been observed infrequently, comprehensive analysis of the frequency and genetic characteristics of such events in the human peripheral blood antibody repertoire has not been possible because of the rarity of such recombinants and the limitations of traditional sequencing technologies. Here, through the use of high-throughput sequencing of the normal human peripheral blood antibody repertoire, we analysed the frequency and genetic characteristics of V(DD)J recombinants. We found that these recombinations were present in approximately 1 in 800 circulating B cells, and that the frequency was severely reduced in memory cell subsets. We also found that V(DD)J recombination can occur across the spectrum of diversity genes, indicating that virtually all recombination signal sequences that flank diversity genes are amenable to V(DD)J recombination. Finally, we observed a repertoire bias in the diversity gene repertoire at the upstream (5') position, and discovered that this bias was primarily attributable to the order of diversity genes in the genomic locus.
© 2012 The Authors. Immunology © 2012 Blackwell Publishing Ltd.
Figures

Similar articles
-
Expressed antibody repertoires in human cord blood cells: 454 sequencing and IMGT/HighV-QUEST analysis of germline gene usage, junctional diversity, and somatic mutations.Immunogenetics. 2012 May;64(5):337-50. doi: 10.1007/s00251-011-0595-8. Epub 2011 Dec 27. Immunogenetics. 2012. PMID: 22200891 Free PMC article.
-
Tissue-specific expressed antibody variable gene repertoires.PLoS One. 2014 Jun 23;9(6):e100839. doi: 10.1371/journal.pone.0100839. eCollection 2014. PLoS One. 2014. PMID: 24956460 Free PMC article.
-
Comprehensive assessment of potential multiple myeloma immunoglobulin heavy chain V-D-J intraclonal variation using massively parallel pyrosequencing.Oncotarget. 2012 Apr;3(4):502-13. doi: 10.18632/oncotarget.469. Oncotarget. 2012. PMID: 22522905 Free PMC article.
-
Diversity in the Cow Ultralong CDR H3 Antibody Repertoire.Front Immunol. 2018 Jun 4;9:1262. doi: 10.3389/fimmu.2018.01262. eCollection 2018. Front Immunol. 2018. PMID: 29915599 Free PMC article. Review.
-
Transcription factories in Igκ allelic choice and diversity.Adv Immunol. 2019;141:33-49. doi: 10.1016/bs.ai.2018.11.001. Epub 2018 Dec 19. Adv Immunol. 2019. PMID: 30904132 Free PMC article. Review.
Cited by
-
Violation of the 12/23 rule of genomic V(D)J recombination is common in lymphocytes.Genome Res. 2015 Feb;25(2):226-34. doi: 10.1101/gr.179770.114. Epub 2014 Nov 3. Genome Res. 2015. PMID: 25367293 Free PMC article.
-
Vaccines combining slow delivery and follicle targeting of antigens increase germinal center B cell clonal diversity and clonal expansion.bioRxiv [Preprint]. 2024 Aug 19:2024.08.19.608655. doi: 10.1101/2024.08.19.608655. bioRxiv. 2024. Update in: Sci Transl Med. 2025 Jun 18;17(803):eadw7499. doi: 10.1126/scitranslmed.adw7499. PMID: 39229011 Free PMC article. Updated. Preprint.
-
Large-scale data mining of four billion human antibody variable regions reveals convergence between therapeutic and natural antibodies that constrains search space for biologics drug discovery.MAbs. 2024 Jan-Dec;16(1):2361928. doi: 10.1080/19420862.2024.2361928. Epub 2024 Jun 6. MAbs. 2024. PMID: 38844871 Free PMC article.
-
Vaccines combining slow release and follicle targeting of antigens increase germinal center B cell diversity and clonal expansion.Sci Transl Med. 2025 Jun 18;17(803):eadw7499. doi: 10.1126/scitranslmed.adw7499. Epub 2025 Jun 18. Sci Transl Med. 2025. PMID: 40531969 Free PMC article.
-
Boosting of HIV envelope CD4 binding site antibodies with long variable heavy third complementarity determining region in the randomized double blind RV305 HIV-1 vaccine trial.PLoS Pathog. 2017 Feb 24;13(2):e1006182. doi: 10.1371/journal.ppat.1006182. eCollection 2017 Feb. PLoS Pathog. 2017. PMID: 28235027 Free PMC article. Clinical Trial.
References
-
- Brack C, Hirama M, Lenhard-Schuller R, Tonegawa S. A complete immunoglobulin gene is created by somatic recombination. Cell. 1978;15:1–14. - PubMed
-
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–81. - PubMed
-
- Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–48. - PubMed
-
- Oettinger M, Schatz D, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–23. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

