Identification of an intracellular M17 family leucine aminopeptidase that is required for virulence in Staphylococcus aureus
- PMID: 22613209
- PMCID: PMC3426635
- DOI: 10.1016/j.micinf.2012.04.013
Identification of an intracellular M17 family leucine aminopeptidase that is required for virulence in Staphylococcus aureus
Abstract
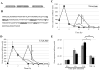
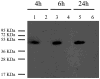
Staphylococcus aureus is a highly virulent bacterial pathogen capable of causing a variety of ailments throughout the human body. It is a major public health concern due to the continued emergence of highly pathogenic methicillin resistant strains (MRSA) both within hospitals and in the community. Virulence in S. aureus is mediated by an array of secreted and cell wall associated virulence factors, including toxins, hemolysins and proteases. In this work we identify a leucine aminopeptidase (LAP, pepZ) that strongly impacts the pathogenic abilities of S. aureus. Disruption of the pepZ gene in either Newman or USA300 resulted in a dramatic attenuation of virulence in both localized and systemic models of infection. LAP is required for survival inside human macrophages and gene expression analysis shows that pepZ expression is highest in the intracellular environment. We examine the cellular location of LAP and demonstrate that it is localized to the bacterial cytosol. These results identify for the first time an intracellular leucine aminopeptidase that influences disease causation in a Gram-positive bacterium.
Copyright © 2012 Institut Pasteur. Published by Elsevier Masson SAS. All rights reserved.
Figures



Similar articles
-
The Staphylococcus aureus leucine aminopeptidase is localized to the bacterial cytosol and demonstrates a broad substrate range that extends beyond leucine.Biol Chem. 2013 Jun;394(6):791-803. doi: 10.1515/hsz-2012-0308. Biol Chem. 2013. PMID: 23241672 Free PMC article.
-
The lone S41 family C-terminal processing protease in Staphylococcus aureus is localized to the cell wall and contributes to virulence.Microbiology (Reading). 2014 Aug;160(Pt 8):1737-1748. doi: 10.1099/mic.0.079798-0. Epub 2014 Jun 13. Microbiology (Reading). 2014. PMID: 24928312 Free PMC article.
-
Simultaneous lack of catalase and beta-toxin in Staphylococcus aureus leads to increased intracellular survival in macrophages and epithelial cells and to attenuated virulence in murine and ovine models.Microbiology (Reading). 2009 May;155(Pt 5):1505-1515. doi: 10.1099/mic.0.025544-0. Epub 2009 Apr 21. Microbiology (Reading). 2009. PMID: 19383704
-
Virulence regulation in Staphylococcus aureus: the need for in vivo analysis of virulence factor regulation.FEMS Immunol Med Microbiol. 2004 Oct 1;42(2):147-54. doi: 10.1016/j.femsim.2004.05.005. FEMS Immunol Med Microbiol. 2004. PMID: 15364098 Review.
-
[Secreted proteins of Staphylococcus aureus].Zh Mikrobiol Epidemiol Immunobiol. 2010 Jul-Aug;(4):118-24. Zh Mikrobiol Epidemiol Immunobiol. 2010. PMID: 20795394 Review. Russian.
Cited by
-
The Staphylococcus aureus leucine aminopeptidase is localized to the bacterial cytosol and demonstrates a broad substrate range that extends beyond leucine.Biol Chem. 2013 Jun;394(6):791-803. doi: 10.1515/hsz-2012-0308. Biol Chem. 2013. PMID: 23241672 Free PMC article.
-
The Small RNA Teg41 Regulates Expression of the Alpha Phenol-Soluble Modulins and Is Required for Virulence in Staphylococcus aureus.mBio. 2019 Feb 5;10(1):e02484-18. doi: 10.1128/mBio.02484-18. mBio. 2019. PMID: 30723124 Free PMC article.
-
The identification of two M20B family peptidases required for full virulence in Staphylococcus aureus.Front Cell Infect Microbiol. 2023 Jul 19;13:1176769. doi: 10.3389/fcimb.2023.1176769. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37538308 Free PMC article.
-
What is behind the epidemiological difference between community-acquired and health-care associated methicillin-resistant Staphylococcus aureus?Virulence. 2017 Aug 18;8(6):640-642. doi: 10.1080/21505594.2017.1335847. Epub 2017 Jun 20. Virulence. 2017. PMID: 28632426 Free PMC article. No abstract available.
-
Proteomics of extracellular vesicles produced by Granulicatella adiacens, which causes infective endocarditis.PLoS One. 2020 Nov 20;15(11):e0227657. doi: 10.1371/journal.pone.0227657. eCollection 2020. PLoS One. 2020. PMID: 33216751 Free PMC article.
References
-
- Gonzales T, Robert-Baudouy J. Bacterial aminopeptidases: properties and functions. FEMS Microbiol Rev. 1996;18:319–344. - PubMed
-
- Christensen JE, Dudley EG, Pederson JA, Steele JL. Peptidases and amino acid catabolism in lactic acid bacteria. Antonie Van Leeuwenhoek. 1999;76:217–246. - PubMed
-
- Savijoki K, Ingmer H, Varmanen P. Proteolytic systems of lactic acid bacteria. Appl Microbiol Biotechnol. 2006;71:394–406. - PubMed
-
- Abid K, Rochat B, Lassahn PG, Stocklin R, Michalet S, Brakch N, Aubert JF, Vatansever B, Tella P, De Meester I, Grouzmann E. Kinetic study of neuropeptide Y (NPY) proteolysis in blood and identification of NPY3-35: a new peptide generated by plasma kallikrein. J Biol Chem. 2009;284:24715–24724. - PMC - PubMed
-
- Barsun M, Jajcanin N, Vukelic B, Spoljaric J, Abramic M. Human dipeptidyl peptidase III acts as a post-proline-cleaving enzyme on endomorphins. Biol Chem. 2007;388:343–348. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

