Resolving the ortholog conjecture: orthologs tend to be weakly, but significantly, more similar in function than paralogs
- PMID: 22615551
- PMCID: PMC3355068
- DOI: 10.1371/journal.pcbi.1002514
Resolving the ortholog conjecture: orthologs tend to be weakly, but significantly, more similar in function than paralogs
Abstract
The function of most proteins is not determined experimentally, but is extrapolated from homologs. According to the "ortholog conjecture", or standard model of phylogenomics, protein function changes rapidly after duplication, leading to paralogs with different functions, while orthologs retain the ancestral function. We report here that a comparison of experimentally supported functional annotations among homologs from 13 genomes mostly supports this model. We show that to analyze GO annotation effectively, several confounding factors need to be controlled: authorship bias, variation of GO term frequency among species, variation of background similarity among species pairs, and propagated annotation bias. After controlling for these biases, we observe that orthologs have generally more similar functional annotations than paralogs. This is especially strong for sub-cellular localization. We observe only a weak decrease in functional similarity with increasing sequence divergence. These findings hold over a large diversity of species; notably orthologs from model organisms such as E. coli, yeast or mouse have conserved function with human proteins.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

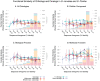


References
-
- Rentzsch R, Orengo CA. Protein function prediction–the power of multiplicity. Trends Biotechnol. 2009;27:210–219. - PubMed
-
- Bork P, Dandekar T, Diaz-Lazcoz Y, Eisenhaber F, Huynen M, et al. Predicting function: from genes to genomes and back. J Mol Biol. 1998;283:707–725. - PubMed
-
- Eisen JA. Phylogenomics: improving functional predictions for uncharacterized genes by evolutionary analysis. Genome Res. 1998;8:163–167. - PubMed
-
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. - PubMed
-
- Studer RA, Robinson-Rechavi M. How confident can we be that orthologs are similar, but paralogs differ? Trends Genet. 2009;25:210–216. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

