Extracellular superoxide dismutase protects Histoplasma yeast cells from host-derived oxidative stress
- PMID: 22615571
- PMCID: PMC3355102
- DOI: 10.1371/journal.ppat.1002713
Extracellular superoxide dismutase protects Histoplasma yeast cells from host-derived oxidative stress
Abstract
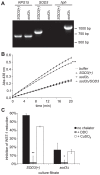
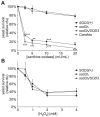
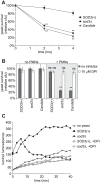
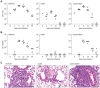
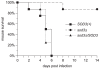
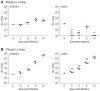
In order to establish infections within the mammalian host, pathogens must protect themselves against toxic reactive oxygen species produced by phagocytes of the immune system. The fungal pathogen Histoplasma capsulatum infects both neutrophils and macrophages but the mechanisms enabling Histoplasma yeasts to survive in these phagocytes have not been fully elucidated. We show that Histoplasma yeasts produce a superoxide dismutase (Sod3) and direct it to the extracellular environment via N-terminal and C-terminal signals which promote its secretion and association with the yeast cell surface. This localization permits Sod3 to protect yeasts specifically from exogenous superoxide whereas amelioration of endogenous reactive oxygen depends on intracellular dismutases such as Sod1. While infection of resting macrophages by Histoplasma does not stimulate the phagocyte oxidative burst, interaction with polymorphonuclear leukocytes (PMNs) and cytokine-activated macrophages triggers production of reactive oxygen species (ROS). Histoplasma yeasts producing Sod3 survive co-incubation with these phagocytes but yeasts lacking Sod3 are rapidly eliminated through oxidative killing similar to the effect of phagocytes on Candida albicans yeasts. The protection provided by Sod3 against host-derived ROS extends in vivo. Without Sod3, Histoplasma yeasts are attenuated in their ability to establish respiratory infections and are rapidly cleared with the onset of adaptive immunity. The virulence of Sod3-deficient yeasts is restored in murine hosts unable to produce superoxide due to loss of the NADPH-oxidase function. These results demonstrate that phagocyte-produced ROS contributes to the immune response to Histoplasma and that Sod3 facilitates Histoplasma pathogenesis by detoxifying host-derived reactive oxygen thereby enabling Histoplasma survival.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Imlay JA. Pathways of oxidative damage. Annu Rev Microbiol. 2003;57:395–418. - PubMed
-
- Johnston RB, Jr, Kitagawa S. Molecular basis for the enhanced respiratory burst of activated macrophages. Fed Proc. 1985;44:2927–2932. - PubMed
-
- Murray HW, Spitalny GL, Nathan CF. Activation of mouse peritoneal macrophages in vitro and in vivo by interferon-gamma. J Immunol. 1985;134:1619–1622. - PubMed
-
- Fleischmann J, Wu-Hsieh B, Howard DH. The intracellular fate of Histoplasma capsulatum in human macrophages is unaffected by recombinant human interferon-gamma. J Infect Dis. 1990;161:143–145. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

