Separation of DNA replication from the assembly of break-competent meiotic chromosomes
- PMID: 22615576
- PMCID: PMC3355065
- DOI: 10.1371/journal.pgen.1002643
Separation of DNA replication from the assembly of break-competent meiotic chromosomes
Abstract
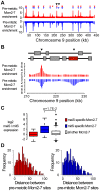
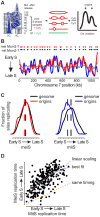
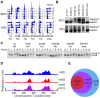
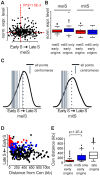
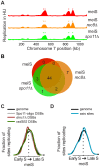
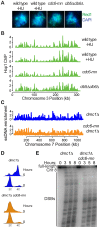
The meiotic cell division reduces the chromosome number from diploid to haploid to form gametes for sexual reproduction. Although much progress has been made in understanding meiotic recombination and the two meiotic divisions, the processes leading up to recombination, including the prolonged pre-meiotic S phase (meiS) and the assembly of meiotic chromosome axes, remain poorly defined. We have used genome-wide approaches in Saccharomyces cerevisiae to measure the kinetics of pre-meiotic DNA replication and to investigate the interdependencies between replication and axis formation. We found that replication initiation was delayed for a large number of origins in meiS compared to mitosis and that meiotic cells were far more sensitive to replication inhibition, most likely due to the starvation conditions required for meiotic induction. Moreover, replication initiation was delayed even in the absence of chromosome axes, indicating replication timing is independent of the process of axis assembly. Finally, we found that cells were able to install axis components and initiate recombination on unreplicated DNA. Thus, although pre-meiotic DNA replication and meiotic chromosome axis formation occur concurrently, they are not strictly coupled. The functional separation of these processes reveals a modular method of building meiotic chromosomes and predicts that any crosstalk between these modules must occur through superimposed regulatory mechanisms.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Comment in
-
Slowing replication in preparation for reduction.PLoS Genet. 2012;8(5):e1002715. doi: 10.1371/journal.pgen.1002715. Epub 2012 May 17. PLoS Genet. 2012. PMID: 22615580 Free PMC article. No abstract available.
References
-
- Moens PB, Pearlman RE. Chromatin organization at meiosis. Bioessays. 1988;9:151–153. - PubMed
-
- Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu Rev Genet. 1999;33:603–754. - PubMed
-
- Williamson DH, Johnston LH, Fennell DJ, Simchen G. The timing of the S phase and other nuclear events in yeast meiosis. Exp Cell Res. 1983;145:209–217. - PubMed
-
- Padmore R, Cao L, Kleckner N. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell. 1991;66:1239–1256. - PubMed
-
- Herrick J. The dynamic replicon: adapting to a changing cellular environment. Bioessays. 2010;32:153–164. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

