Phylogenomics of prokaryotic ribosomal proteins
- PMID: 22615861
- PMCID: PMC3353972
- DOI: 10.1371/journal.pone.0036972
Phylogenomics of prokaryotic ribosomal proteins
Abstract
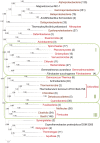
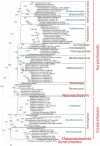
Archaeal and bacterial ribosomes contain more than 50 proteins, including 34 that are universally conserved in the three domains of cellular life (bacteria, archaea, and eukaryotes). Despite the high sequence conservation, annotation of ribosomal (r-) protein genes is often difficult because of their short lengths and biased sequence composition. We developed an automated computational pipeline for identification of r-protein genes and applied it to 995 completely sequenced bacterial and 87 archaeal genomes available in the RefSeq database. The pipeline employs curated seed alignments of r-proteins to run position-specific scoring matrix (PSSM)-based BLAST searches against six-frame genome translations, mitigating possible gene annotation errors. As a result of this analysis, we performed a census of prokaryotic r-protein complements, enumerated missing and paralogous r-proteins, and analyzed the distributions of ribosomal protein genes among chromosomal partitions. Phyletic patterns of bacterial and archaeal r-protein genes were mapped to phylogenetic trees reconstructed from concatenated alignments of r-proteins to reveal the history of likely multiple independent gains and losses. These alignments, available for download, can be used as search profiles to improve genome annotation of r-proteins and for further comparative genomics studies.
Conflict of interest statement
Figures





References
-
- Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell. 2002;108:557–572. - PubMed
-
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. - PubMed
-
- Wimberly BT, Brodersen DE, Clemons WM, Morgan-Warren RJ, Carter AP, et al. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. - PubMed
-
- Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, et al. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 2000;102:615–623. - PubMed
-
- Williamson JR. The ribosome at atomic resolution. Cell. 2009;139:1041–1043. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

