Highly sensitive in vitro methods for detection of residual undifferentiated cells in retinal pigment epithelial cells derived from human iPS cells
- PMID: 22615985
- PMCID: PMC3355139
- DOI: 10.1371/journal.pone.0037342
Highly sensitive in vitro methods for detection of residual undifferentiated cells in retinal pigment epithelial cells derived from human iPS cells
Abstract
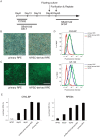
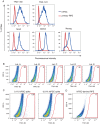
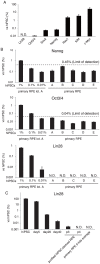
Human induced pluripotent stem cells (hiPSCs) possess the capabilities of self-renewal and differentiation into multiple cell types, and they are free of the ethical problems associated with human embryonic stem cells (hESCs). These characteristics make hiPSCs a promising choice for future regenerative medicine research. There are significant obstacles, however, preventing the clinical use of hiPSCs. One of the most obvious safety issues is the presence of residual undifferentiated cells that have tumorigenic potential. To locate residual undifferentiated cells, in vivo teratoma formation assays have been performed with immunodeficient animals, which is both costly and time-consuming. Here, we examined three in vitro assay methods to detect undifferentiated cells (designated an in vitro tumorigenicity assay): soft agar colony formation assay, flow cytometry assay and quantitative real-time polymerase chain reaction assay (qRT-PCR). Although the soft agar colony formation assay was unable to detect hiPSCs even in the presence of a ROCK inhibitor that permits survival of dissociated hiPSCs/hESCs, the flow cytometry assay using anti-TRA-1-60 antibody detected 0.1% undifferentiated hiPSCs that were spiked in primary retinal pigment epithelial (RPE) cells. Moreover, qRT-PCR with a specific probe and primers was found to detect a trace amount of Lin28 mRNA, which is equivalent to that present in a mixture of a single hiPSC and 5.0×10⁴ RPE cells. Our findings provide highly sensitive and quantitative in vitro assays essential for facilitating safety profiling of hiPSC-derived products for future regenerative medicine research.
Conflict of interest statement
Figures

References
-
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. - PubMed
-
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. - PubMed
-
- Zhang SC, Wernig M, Duncan ID, Brustle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19:1129–1133. - PubMed
-
- Cai J, Zhao Y, Liu Y, Ye F, Song Z, et al. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology. 2007;45:1229–1239. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

