Cryptic transcripts from a ubiquitous plasmid origin of replication confound tests for cis-regulatory function
- PMID: 22618870
- PMCID: PMC3424574
- DOI: 10.1093/nar/gks451
Cryptic transcripts from a ubiquitous plasmid origin of replication confound tests for cis-regulatory function
Abstract
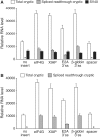
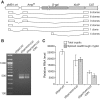
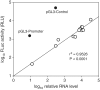
A vast amount of research on the regulation of gene expression has relied on plasmid reporter assays. In this study, we show that plasmids widely used for this purpose constitutively produce substantial amounts of RNA from a TATA-containing cryptic promoter within the origin of replication. Readthrough of these RNAs into the intended transcriptional unit potently stimulated reporter activity when the inserted test sequence contained a 3' splice site (ss). We show that two human sequences, originally reported to be internal ribosome entry sites and later to instead be promoters, mimic both types of element in dicistronic reporter assays by causing these cryptic readthrough transcripts to splice in patterns that allow efficient translation of the downstream cistron. Introduction of test sequences containing 3' ss into monocistronic luciferase reporter vectors widely used in the study of transcriptional regulation also created the false appearance of promoter function via the same mechanism. Across a large number of variants of these plasmids, we found a very highly significant correlation between reporter activity and levels of such spliced readthrough transcripts. Computational estimation of the frequency of cryptic 3' ss in genomic sequences suggests that misattribution of cis-regulatory function may be a common occurrence.
Figures





References
-
- Carey M, Peterson CL, Smale ST. Transcriptional Regulation in Eukaryotes: Concepts, Strategies, and Techniques. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2009.
-
- Maston GA, Evans SK, Green MR. Transcriptional regulatory elements in the human genome. Annu. Rev. Genomics Hum. Genet. 2006;7:29–59. - PubMed
-
- Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

