JMJD6 is a driver of cellular proliferation and motility and a marker of poor prognosis in breast cancer
- PMID: 22621393
- PMCID: PMC3446348
- DOI: 10.1186/bcr3200
JMJD6 is a driver of cellular proliferation and motility and a marker of poor prognosis in breast cancer
Erratum in
-
Erratum to: JMJD6 is a driver of cellular proliferation and motility and a marker of poor prognosis in breast cancer.Breast Cancer Res. 2017 Mar 28;19(1):42. doi: 10.1186/s13058-017-0830-9. Breast Cancer Res. 2017. PMID: 28351428 Free PMC article. No abstract available.
Abstract
Introduction: We developed an analytic strategy that correlates gene expression and clinical outcomes as a means to identify novel candidate oncogenes operative in breast cancer. This analysis, followed by functional characterization, resulted in the identification of Jumonji Domain Containing 6 (JMJD6) protein as a novel driver of oncogenic properties in breast cancer.
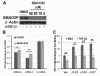
Methods: Through microarray informatics, Cox proportional hazards regression was used to analyze the correlation between gene expression and distant metastasis-free survival (DMFS) of patients in 14 independent breast cancer cohorts. JMJD6 emerged as a top candidate gene robustly associated with poor patient survival. Immunohistochemistry, siRNA-mediated silencing, and forced overexpression of JMJD6 in cell-based assays elucidated molecular mechanisms of JMJD6 action in breast cancer progression and shed light on the clinical breast cancer subtypes relevant to JMJD6 action.
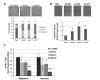
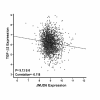
Results: JMJD6 was expressed at highest levels in tumors associated with worse outcomes, including ER- and basal-like, Claudin-low, Her2-enriched, and ER+ Luminal B tumors. High nuclear JMJD6 protein was associated with ER negativity, advanced grade, and poor differentiation in tissue microarrays. Separation of ER+/LN- patients that received endocrine monotherapy indicated that JMJD6 is predictive of poor outcome in treatment-specific subgroups. In breast cancer cell lines, loss of JMJD6 consistently resulted in suppressed proliferation but not apoptosis, whereas forced stable overexpression increased growth. In addition, knockdown of JMJD6 in invasive cell lines, such as MDA-MB231, decreased motility and invasion, whereas overexpression in MCF-7 cells slightly promoted motility but did not confer invasive growth. Microarray analysis showed that the most significant transcriptional changes occurred in cell-proliferation genes and genes of the TGF-β tumor-suppressor pathway. High proliferation was characterized by constitutively high cyclin E protein levels. The inverse relation of JMJD6 expression with TGF-β2 could be extrapolated to the breast cancer cohorts, suggesting that JMJD6 may affect similar pathways in primary breast cancer.
Conclusions: JMJD6 is a novel biomarker of tumor aggressiveness with functional implications in breast cancer growth and migration.
Figures

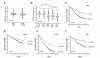

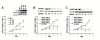




References
-
- Soon WWML, Black MA, Dalmasso C, Chan XB, Pang B, Ong CW, Salto-Tellez M, Desai K, Liu ET. Combined genomic and phenotype screening reveals secretory factor SPINK1 as an invasion and survival factor associated with patient prognosis in breast cancer. EMBO Mol Med. 2011;3:451–464. doi: 10.1002/emmm.201100150. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

