Lineage-specific adjacent IFNG and IL26 genes share a common distal enhancer element
- PMID: 22622197
- PMCID: PMC4180225
- DOI: 10.1038/gene.2012.22
Lineage-specific adjacent IFNG and IL26 genes share a common distal enhancer element
Abstract
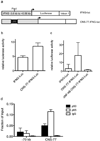
Certain groups of physically linked genes remain linked over long periods of evolutionary time. The general view is that such evolutionary conservation confers 'fitness' to the species. Why gene order confers 'fitness' to the species is incompletely understood. For example, linkage of IL26 and IFNG is preserved over evolutionary time yet Th17 lineages express IL26 and Th1 lineages express IFNG. We considered the hypothesis that distal enhancer elements may be shared between adjacent genes, which would require linkage be maintained in evolution. We test this hypothesis using a bacterial artificial chromosome transgenic model with deletions of specific conserved non-coding sequences. We identify one enhancer element uniquely required for IL26 expression but not for IFNG expression. We identify a second enhancer element positioned between IL26 and IFNG required for both IL26 and IFNG expression. One function of this enhancer is to facilitate recruitment of RNA polymerase II to promoters of both genes. Thus, sharing of distal enhancers between adjacent genes may contribute to evolutionary preservation of gene order.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






References
-
- Boniface K, Blumenschein WM, Brovont-Porth K, McGeachy MJ, Basham B, Desai B, et al. Human Th17 cells comprise heterogeneous subsets including IFN-gamma-producing cells with distinct properties from the Th1 lineage. J Immunol. 2010;185(1):679–687. - PubMed
-
- Commins S, Steinke JW, Borish L. The extended IL-10 superfamily: IL-10, IL-19, IL-20, IL-22, IL-24, IL-26, IL-28, and IL-29. J Allergy Clin Immunol. 2008;121(5):1108–1111. - PubMed
-
- Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

