Peroxiredoxins are conserved markers of circadian rhythms
- PMID: 22622569
- PMCID: PMC3398137
- DOI: 10.1038/nature11088
Peroxiredoxins are conserved markers of circadian rhythms
Erratum in
- Nature. 2012 Sep 27;489(7417):590
Abstract
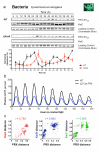
Cellular life emerged ∼3.7 billion years ago. With scant exception, terrestrial organisms have evolved under predictable daily cycles owing to the Earth's rotation. The advantage conferred on organisms that anticipate such environmental cycles has driven the evolution of endogenous circadian rhythms that tune internal physiology to external conditions. The molecular phylogeny of mechanisms driving these rhythms has been difficult to dissect because identified clock genes and proteins are not conserved across the domains of life: Bacteria, Archaea and Eukaryota. Here we show that oxidation-reduction cycles of peroxiredoxin proteins constitute a universal marker for circadian rhythms in all domains of life, by characterizing their oscillations in a variety of model organisms. Furthermore, we explore the interconnectivity between these metabolic cycles and transcription-translation feedback loops of the clockwork in each system. Our results suggest an intimate co-evolution of cellular timekeeping with redox homeostatic mechanisms after the Great Oxidation Event ∼2.5 billion years ago.
Figures





References
-
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. - PubMed
-
- Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks; an experimental assessment in cyanobacteria. Curr Biol. 2004;14:1481–6. - PubMed
-
- Dodd AN, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–3. - PubMed
-
- Barger LK, Lockley SW, Rajaratnam SM, Landrigan CP. Neurobehavioral, health, and safety consequences associated with shift work in safety-sensitive professions. Curr Neurol Neurosci Rep. 2009;9:155–64. - PubMed
-
- Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Annu Rev Genet. 2006;40:409–48. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50GM076547/GM/NIGMS NIH HHS/United States
- 093734/Z/10/Z/WT_/Wellcome Trust/United Kingdom
- R21 HL102492/HL/NHLBI NIH HHS/United States
- MC_UP_1201/4/MRC_/Medical Research Council/United Kingdom
- MC_U105170643/MRC_/Medical Research Council/United Kingdom
- 083643/Z/07/Z/WT_/Wellcome Trust/United Kingdom
- BB/D019621/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01GM088595/GM/NIGMS NIH HHS/United States
- P50 GM076547/GM/NIGMS NIH HHS/United States
- BB/C006941/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G0600717/MRC_/Medical Research Council/United Kingdom
- R37 GM067152/GM/NIGMS NIH HHS/United States
- 093734/WT_/Wellcome Trust/United Kingdom
- R01 GM088595/GM/NIGMS NIH HHS/United States
- BB/D019621/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R21HL102492/HL/NHLBI NIH HHS/United States
- R01 GM067152/GM/NIGMS NIH HHS/United States
- 083643/WT_/Wellcome Trust/United Kingdom
- R01GM067152/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

