Melanoma genome sequencing reveals frequent PREX2 mutations
- PMID: 22622578
- PMCID: PMC3367798
- DOI: 10.1038/nature11071
Melanoma genome sequencing reveals frequent PREX2 mutations
Abstract
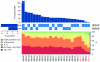
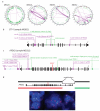
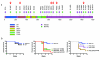
Melanoma is notable for its metastatic propensity, lethality in the advanced setting and association with ultraviolet exposure early in life. To obtain a comprehensive genomic view of melanoma in humans, we sequenced the genomes of 25 metastatic melanomas and matched germline DNA. A wide range of point mutation rates was observed: lowest in melanomas whose primaries arose on non-ultraviolet-exposed hairless skin of the extremities (3 and 14 per megabase (Mb) of genome), intermediate in those originating from hair-bearing skin of the trunk (5-55 per Mb), and highest in a patient with a documented history of chronic sun exposure (111 per Mb). Analysis of whole-genome sequence data identified PREX2 (phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2)--a PTEN-interacting protein and negative regulator of PTEN in breast cancer--as a significantly mutated gene with a mutation frequency of approximately 14% in an independent extension cohort of 107 human melanomas. PREX2 mutations are biologically relevant, as ectopic expression of mutant PREX2 accelerated tumour formation of immortalized human melanocytes in vivo. Thus, whole-genome sequencing of human melanoma tumours revealed genomic evidence of ultraviolet pathogenesis and discovered a new recurrently mutated gene in melanoma.
Figures
Comment in
-
Registered report: Melanoma genome sequencing reveals frequent PREX2 mutations.Elife. 2014 Dec 10;3:e04180. doi: 10.7554/eLife.04180. Elife. 2014. PMID: 25490935 Free PMC article.
References
-
- Chin L. The genetics of malignant melanoma: lessons from mouse and man. Nat Rev Cancer. 2003;3:559–70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

