Workflow for analysis of high mass accuracy salivary data set using MaxQuant and ProteinPilot search algorithm
- PMID: 22623410
- PMCID: PMC3618284
- DOI: 10.1002/pmic.201100097
Workflow for analysis of high mass accuracy salivary data set using MaxQuant and ProteinPilot search algorithm
Abstract
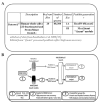
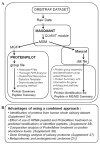
LTQ Orbitrap data analyzed with ProteinPilot can be further improved by MaxQuant raw data processing, which utilizes precursor-level high mass accuracy data for peak processing and MGF creation. In particular, ProteinPilot results from MaxQuant-processed peaklists for Orbitrap data sets resulted in improved spectral utilization due to an improved peaklist quality with higher precision and high precursor mass accuracy (HPMA). The output and postsearch analysis tools of both workflows were utilized for previously unexplored features of a three-dimensional fractionated and hexapeptide library (ProteoMiner) treated whole saliva data set comprising 200 fractions. ProteinPilot's ability to simultaneously predict multiple modifications showed an advantage from ProteoMiner treatment for modified peptide identification. We demonstrate that complementary approaches in the analysis pipeline provide comprehensive results for the whole saliva data set acquired on an LTQ Orbitrap. Overall our results establish a workflow for improved protein identification from high mass accuracy data.
© 2012 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
The authors have declared no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

