Integrative genomic analysis implicates gain of PIK3CA at 3q26 and MYC at 8q24 in chronic lymphocytic leukemia
- PMID: 22623730
- PMCID: PMC3719990
- DOI: 10.1158/1078-0432.CCR-11-2342
Integrative genomic analysis implicates gain of PIK3CA at 3q26 and MYC at 8q24 in chronic lymphocytic leukemia
Abstract
Purpose: The disease course of chronic lymphocytic leukemia (CLL) varies significantly within cytogenetic groups. We hypothesized that high-resolution genomic analysis of CLL would identify additional recurrent abnormalities associated with short time-to-first therapy (TTFT).
Experimental design: We undertook high-resolution genomic analysis of 161 prospectively enrolled CLLs using Affymetrix 6.0 SNP arrays, and integrated analysis of this data set with gene expression profiles.
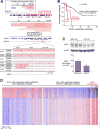
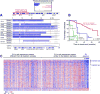
Results: Copy number analysis (CNA) of nonprogressive CLL reveals a stable genotype, with a median of only 1 somatic CNA per sample. Progressive CLL with 13q deletion was associated with additional somatic CNAs, and a greater number of CNAs was predictive of TTFT. We identified other recurrent CNAs associated with short TTFT: 8q24 amplification focused on the cancer susceptibility locus near MYC in 3.7%; 3q26 amplifications focused on PIK3CA in 5.6%; and 8p deletions in 5% of patients. Sequencing of MYC further identified somatic mutations in two CLLs. We determined which catalytic subunits of phosphoinositide 3-kinase (PI3K) were in active complex with the p85 regulatory subunit and showed enrichment for the α subunit in three CLLs carrying PIK3CA amplification.
Conclusions: Our findings implicate amplifications of 3q26 focused on PIK3CA and 8q24 focused on MYC in CLL.
Conflict of interest statement
Figures



References
-
- Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. The New England journal of medicine. 2000;343:1910–6. - PubMed
-
- Grubor V, Krasnitz A, Troge JE, Meth JL, Lakshmi B, Kendall JT, et al. Novel genomic alterations and clonal evolution in chronic lymphocytic leukemia revealed by representational oligonucleotide microarray analysis (ROMA) Blood. 2009;113:1294–303. - PubMed
-
- Gunnarsson R, Isaksson A, Mansouri M, Goransson H, Jansson M, Cahill N, et al. Large but not small copy-number alterations correlate to high-risk genomic aberrations and survival in chronic lymphocytic leukemia: a high-resolution genomic screening of newly diagnosed patients. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2010;24:211–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

