Cellular GCN5 is a novel regulator of human adenovirus E1A-conserved region 3 transactivation
- PMID: 22623781
- PMCID: PMC3421684
- DOI: 10.1128/JVI.00289-12
Cellular GCN5 is a novel regulator of human adenovirus E1A-conserved region 3 transactivation
Abstract
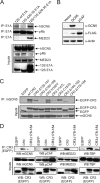
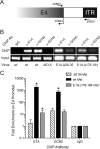
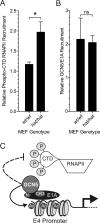
The largest isoform of adenovirus early region 1A (E1A) contains a unique region termed conserved region 3 (CR3). This region activates viral gene expression by recruiting cellular transcription machinery to the early viral promoters. Recent studies have suggested that there is an optimal level of E1A-dependent transactivation required by human adenovirus (hAd) during infection and that this may be achieved via functional cross talk between the N termini of E1A and CR3. The N terminus of E1A binds GCN5, a cellular lysine acetyltransferase (KAT). We have identified a second independent interaction of E1A with GCN5 that is mediated by CR3, which requires residues 178 to 188 in hAd5 E1A. GCN5 was recruited to the viral genome during infection in an E1A-dependent manner, and this required both GCN5 interaction sites on E1A. Ectopic expression of GCN5 repressed transactivation by both E1A CR3 and full-length E1A. In contrast, RNA interference (RNAi) depletion of GCN5 or treatment with the KAT inhibitor cyclopentylidene-[4-(4'-chlorophenyl)thiazol-2-yl]hydrazone (CPTH2) resulted in increased E1A CR3 transactivation. Moreover, activation of the adenovirus E4 promoter by E1A was increased during infection of homozygous GCN5 KAT-defective (hat/hat) mouse embryonic fibroblasts (MEFs) compared to wild-type control MEFs. Enhanced histone H3 K9/K14 acetylation at the viral E4 promoter required the newly identified binding site for GCN5 within CR3 and correlated with repression and reduced occupancy by phosphorylated RNA polymerase II. Treatment with CPTH2 during infection also reduced virus yield. These data identify GCN5 as a new negative regulator of transactivation by E1A and suggest that its KAT activity is required for optimal virus replication.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

