New PARP gene with an anti-alphavirus function
- PMID: 22623789
- PMCID: PMC3421642
- DOI: 10.1128/JVI.00733-12
New PARP gene with an anti-alphavirus function
Abstract
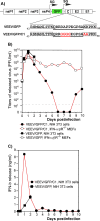
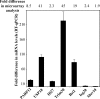
Alphaviruses represent a highly important group of human and animal pathogens, which are transmitted by mosquito vectors between vertebrate hosts. The hallmark of alphavirus infection in vertebrates is the induction of a high-titer viremia, which is strongly dependent on the ability of the virus to interfere with host antiviral responses on both cellular and organismal levels. The identification of cellular factors, which are critical in orchestrating virus clearance without the development of cytopathic effect, may prove crucial in the design of new and highly effective antiviral treatments. To address this issue, we have developed a noncytopathic Venezuelan equine encephalitis virus (VEEV) mutant that can persistently replicate in cells defective in type I interferon (IFN) production or signaling but is cleared from IFN signaling-competent cells. Using this mutant, we analyzed (i) the spectrum of cellular genes activated by virus replication in the persistently infected cells and (ii) the spectrum of genes activated during noncytopathic virus clearance. By applying microarray-based technology and bioinformatic analysis, we identified a number of IFN-stimulated genes (ISGs) specifically activated during VEEV clearance. One of these gene products, the long isoform of PARP12 (PARP12L), demonstrated an inhibitory effect on the replication of VEEV, as well as other alphaviruses and several different types of other RNA viruses. Additionally, overexpression of two other members of the PARP gene superfamily was also shown to be capable of inhibiting VEEV replication.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

