Fluorescence correlation spectroscopy in drug discovery: study of Alexa532-endothelin 1 binding to the endothelin ETA receptor to describe the pharmacological profile of natural products
- PMID: 22623909
- PMCID: PMC3353486
- DOI: 10.1100/2012/524169
Fluorescence correlation spectroscopy in drug discovery: study of Alexa532-endothelin 1 binding to the endothelin ETA receptor to describe the pharmacological profile of natural products
Abstract
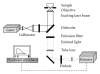
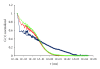
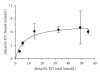
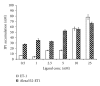
Fluorescence correlation spectroscopy and the newly synthesized Alexa532-ET1 were used to study the dynamics of the endothelin ET(A) receptor-ligand complex alone and under the influence of a semisynthetic selective antagonist and a fungal extract on living A10 cells. Dose-dependent increase of inositol phosphate production was seen for Alexa532-ET1, and its binding was reduced to 8% by the selective endothelin ET(A) antagonist BQ-123, confirming the specific binding of Alexa532-ET1 to the endothelin ET(A) receptor. Two different lateral mobilities of the receptor-ligand complexes within the cell membrane were found allowing the discrimination of different states for this complex. BQ-123 showed a strong binding affinity to the "inactive" receptor state characterized by the slow diffusion time constant. A similar effect was observed for the fungal extract, which completely displaced Alexa532-ET1 from its binding to the "inactive" receptor state. These findings suggest that both BQ-123 and the fungal extract act as inverse agonists.
Figures
References
-
- Glaser KB, Mayer AMS. A renaissance in marine pharmacology: from preclinical curiosity to clinical reality. Biochemical Pharmacology. 2009;78(5):440–448. - PubMed
-
- Molinski TF, Dalisay DS, Lievens SL, Saludes JP. Drug development from marine natural products. Nature Reviews Drug Discovery. 2009;8(1):69–85. - PubMed
-
- Yanagisawa M, Kurihara H, Kimura S, et al. A novel potent vasoconstrictor peptide produced by vascular endothelial cells. Nature. 1988;332(6163):411–415. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

