Mitochondrial and plasma membrane pools of stomatin-like protein 2 coalesce at the immunological synapse during T cell activation
- PMID: 22623988
- PMCID: PMC3356372
- DOI: 10.1371/journal.pone.0037144
Mitochondrial and plasma membrane pools of stomatin-like protein 2 coalesce at the immunological synapse during T cell activation
Abstract
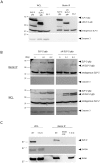
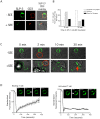
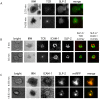
Stomatin-like protein 2 (SLP-2) is a member of the stomatin-prohibitin-flotillin-HflC/K (SPFH) superfamily. Recent evidence indicates that SLP-2 is involved in the organization of cardiolipin-enriched microdomains in mitochondrial membranes and the regulation of mitochondrial biogenesis and function. In T cells, this role translates into enhanced T cell activation. Although the major pool of SLP-2 is associated with mitochondria, we show here that there is an additional pool of SLP-2 associated with the plasma membrane of T cells. Both plasma membrane-associated and mitochondria-associated pools of SLP-2 coalesce at the immunological synapse (IS) upon T cell activation. SLP-2 is not required for formation of IS nor for the re-localization of mitochondria to the IS because SLP-2-deficient T cells showed normal re-localization of these organelles in response to T cell activation. Interestingly, upon T cell activation, we found the surface pool of SLP-2 mostly excluded from the central supramolecular activation complex, and enriched in the peripheral area of the IS where signalling TCR microclusters are located. Based on these results, we propose that SLP-2 facilitates the compartmentalization not only of mitochondrial membranes but also of the plasma membrane into functional microdomains. In this latter location, SLP-2 may facilitate the optimal assembly of TCR signalosome components. Our data also suggest that there may be a net exchange of membrane material between mitochondria and plasma membrane, explaining the presence of some mitochondrial proteins in the plasma membrane.
Conflict of interest statement
Figures

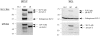

References
-
- Da Cruz S, Xenarios I, Langridge J, Vilbois F, Parone PA, et al. Proteomic analysis of the mouse liver mitochondrial inner membrane. J Biol Chem. 2003;278:41566–41571. - PubMed
-
- Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, et al. Characterization of the human heart mitochondrial proteome. Nat Biotechnol. 2003;21:281–286. - PubMed
-
- Hajek P, Chomyn A, Attardi G. Identification of a novel mitochondrial complex containing mitofusin 2 and stomatin-like protein 2. J Biol Chem. 2007;282:5670–5681. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

