Allele-specific p53 mutant reactivation
- PMID: 22624712
- PMCID: PMC3366694
- DOI: 10.1016/j.ccr.2012.03.042
Allele-specific p53 mutant reactivation
Abstract
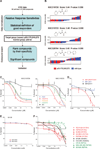
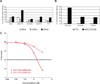
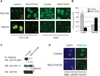
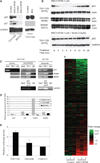
Rescuing the function of mutant p53 protein is an attractive cancer therapeutic strategy. Using the National Cancer Institute's anticancer drug screen data, we identified two compounds from the thiosemicarbazone family that manifest increased growth inhibitory activity in mutant p53 cells, particularly for the p53(R175) mutant. Mechanistic studies reveal that NSC319726 restores WT structure and function to the p53(R175) mutant. This compound kills p53(R172H) knockin mice with extensive apoptosis and inhibits xenograft tumor growth in a 175-allele-specific mutant p53-dependent manner. This activity depends upon the zinc ion chelating properties of the compound as well as redox changes. These data identify NSC319726 as a p53(R175) mutant reactivator and as a lead compound for p53-targeted drug development.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Anticancer drugs: Reactivating p53.Nat Rev Drug Discov. 2012 Jun 29;11(7):517. doi: 10.1038/nrd3782. Nat Rev Drug Discov. 2012. PMID: 22743974 No abstract available.
Similar articles
-
Reactivation of mutant p53 by a dietary-related compound phenethyl isothiocyanate inhibits tumor growth.Cell Death Differ. 2016 Oct;23(10):1615-27. doi: 10.1038/cdd.2016.48. Epub 2016 Jun 3. Cell Death Differ. 2016. PMID: 27258787 Free PMC article.
-
COTI-2 reactivates mutant p53 and inhibits growth of triple-negative breast cancer cells.Breast Cancer Res Treat. 2020 Jan;179(1):47-56. doi: 10.1007/s10549-019-05435-1. Epub 2019 Sep 19. Breast Cancer Res Treat. 2020. PMID: 31538264
-
A novel small molecule FL118 that selectively inhibits survivin, Mcl-1, XIAP and cIAP2 in a p53-independent manner, shows superior antitumor activity.PLoS One. 2012;7(9):e45571. doi: 10.1371/journal.pone.0045571. Epub 2012 Sep 19. PLoS One. 2012. PMID: 23029106 Free PMC article.
-
Zinc binding and redox control of p53 structure and function.Antioxid Redox Signal. 2001 Aug;3(4):611-23. doi: 10.1089/15230860152542961. Antioxid Redox Signal. 2001. PMID: 11554448 Review.
-
Reactivating mutant p53 using small molecules as zinc metallochaperones: awakening a sleeping giant in cancer.Drug Discov Today. 2015 Nov;20(11):1391-7. doi: 10.1016/j.drudis.2015.07.006. Epub 2015 Jul 20. Drug Discov Today. 2015. PMID: 26205328 Free PMC article. Review.
Cited by
-
Current insights into the regulation of programmed cell death by TP53 mutation in cancer.Front Oncol. 2022 Oct 13;12:1023427. doi: 10.3389/fonc.2022.1023427. eCollection 2022. Front Oncol. 2022. PMID: 36313700 Free PMC article. Review.
-
TP53: an oncogene in disguise.Cell Death Differ. 2015 Aug;22(8):1239-49. doi: 10.1038/cdd.2015.53. Epub 2015 May 29. Cell Death Differ. 2015. PMID: 26024390 Free PMC article. Review.
-
Mouse p53-deficient cancer models as platforms for obtaining genomic predictors of human cancer clinical outcomes.PLoS One. 2012;7(8):e42494. doi: 10.1371/journal.pone.0042494. Epub 2012 Aug 7. PLoS One. 2012. PMID: 22880004 Free PMC article.
-
Therapeutic targeting of the p53 pathway in cancer stem cells.Expert Opin Ther Targets. 2012 Dec;16(12):1161-74. doi: 10.1517/14728222.2012.726985. Epub 2012 Sep 24. Expert Opin Ther Targets. 2012. PMID: 22998602 Free PMC article. Review.
-
TCRP1 activated by mutant p53 promotes NSCLC proliferation via inhibiting FOXO3a.Oncogenesis. 2022 Apr 22;11(1):19. doi: 10.1038/s41389-022-00392-9. Oncogenesis. 2022. PMID: 35459265 Free PMC article.
References
-
- Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, Solari A, Bobisse S, Rondina MB, Guzzardo V, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. - PubMed
-
- Butler JS, Loh SN. Structure, function, and aggregation of the zinc-free form of the p53 DNA binding domain. Biochemistry. 2003;42:2396–2403. - PubMed
-
- Butler JS, Loh SN. Zn(2+)-dependent misfolding of the p53 DNA binding domain. Biochemistry. 2007;46:2630–2639. - PubMed
-
- Bykov VJ, Issaeva N, Shilov A, Hultcrantz M, Pugacheva E, Chumakov P, Bergman J, Wiman KG, Selivanova G. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med. 2002;8:282–288. - PubMed
-
- Bykov VJ, Issaeva N, Zache N, Shilov A, Hultcrantz M, Bergman J, Selivanova G, Wiman KG. Reactivation of mutant p53 and induction of apoptosis in human tumor cells by maleimide analogs. J Biol Chem. 2005;280:30384–30391. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

