In vivo imaging of tumor-propagating cells, regional tumor heterogeneity, and dynamic cell movements in embryonal rhabdomyosarcoma
- PMID: 22624717
- PMCID: PMC3381357
- DOI: 10.1016/j.ccr.2012.03.043
In vivo imaging of tumor-propagating cells, regional tumor heterogeneity, and dynamic cell movements in embryonal rhabdomyosarcoma
Abstract
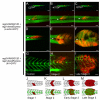
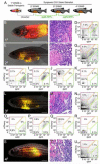
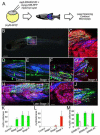
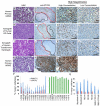
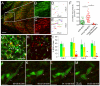
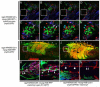
Embryonal rhabdomyosarcoma (ERMS) is an aggressive pediatric sarcoma of muscle. Here, we show that ERMS-propagating potential is confined to myf5+ cells and can be visualized in live, fluorescent transgenic zebrafish. During early tumor growth, myf5+ ERMS cells reside adjacent normal muscle fibers. By late-stage ERMS, myf5+ cells are reorganized into distinct regions separated from differentiated tumor cells. Time-lapse imaging of late-stage ERMS revealed that myf5+ cells populate newly formed tumor only after seeding by highly migratory myogenin+ ERMS cells. Moreover, myogenin+ ERMS cells can enter the vasculature, whereas myf5+ ERMS-propagating cells do not. Our data suggest that non-tumor-propagating cells likely have important supportive roles in cancer progression and facilitate metastasis.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures

References
-
- Conboy IM, Conboy MJ, Smythe GM, Rando TA. Notch-mediated restoration of regenerative potential to aged muscle. Science. 2003;302:1575–1577. - PubMed
-
- Cooper RN, Tajbakhsh S, Mouly V, Cossu G, Buckingham M, Butler-Browne GS. In vivo satellite cell activation via Myf5 and MyoD in regenerating mouse skeletal muscle. J Cell Sci. 1999;112(Pt 17):2895–2901. - PubMed
-
- Dalerba P, Cho RW, Clarke MF. Cancer stem cells: models and concepts. Annu Rev Med. 2007;58:267–284. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- R24 OD016761/OD/NIH HHS/United States
- R01 CA122706/CA/NCI NIH HHS/United States
- R01 CA154923/CA/NCI NIH HHS/United States
- T32 HL007627/HL/NHLBI NIH HHS/United States
- T32 CA009216/CA/NCI NIH HHS/United States
- K12 HD043494/HD/NICHD NIH HHS/United States
- R01 CA150975/CA/NCI NIH HHS/United States
- 5T32CA09216-26/CA/NCI NIH HHS/United States
- R21 CA156056/CA/NCI NIH HHS/United States
- 1R01CA154923/CA/NCI NIH HHS/United States
- K08 AR063165/AR/NIAMS NIH HHS/United States
- 1R21CA156056/CA/NCI NIH HHS/United States
- K01 AR055619/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

