Identification of novel low molecular weight serum peptidome biomarkers for non-small cell lung cancer (NSCLC)
- PMID: 22628229
- PMCID: PMC6807414
- DOI: 10.1002/jcla.21502
Identification of novel low molecular weight serum peptidome biomarkers for non-small cell lung cancer (NSCLC)
Abstract
Aim: To identify discriminating protein patterns in serum samples among non-small cell lung cancer (NSCLC), chronic obstructive pulmonary disease (COPD), pneumonia, and healthy controls. To discover specific low molecular weight (LMW) serum peptidome biomarkers and establish a diagnostic pattern for NSCLCby using proteomic technology.
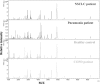
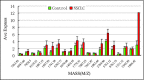
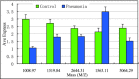
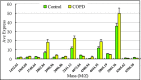
Methods: We used magnetic bead-based separation followed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) to identify patients with NSCLC, COPD, and pneumonia. A total of 154 serum samples were analyzed in this study, among which there were 60 serum samples from NSCLC patients, 30 from patients with other lung-related diseases (16 pneumonia patients and 14 patients with COPD) as disease controls, and 64 from healthy volunteers as healthy control. The mass spectra, analyzed using ClinProTools software, distinguished between cancer patients and healthy individuals based on GA algorithm model.
Results: In this study, we generated numerous discriminating m/z peaks as well as disease-specific discrimination peaks. A set of five potential biomarkers (m/z: 7,763.24, 1,012.61, 4,153.16, 1,450.55, and 2,878.89) could be used as the diagnostic biomarkers to distinguish NSCLCpatients from healthy controls. In the training set, patients with NSCLC could be identified with sensitivity of 97.5% and specificity of 98.8%. Similar results were obtained in the testing set, showing 80.7% sensitivity and 91.2% specificity.
Conclusion: Our study demonstrated that a combined application of magnetic beads with MALDI-TOF MS technique was suitable for identification of serum biomarkers for NSCLC.
© 2012 Wiley Periodicals, Inc.
Conflict of interest statement
The authors confirm that they have no financial or personal relationships that cause a conflict of interest regarding the work in the manuscript.
Figures
Similar articles
-
Classification of Epidermal Growth Factor Receptor Gene Mutation Status Using Serum Proteomic Profiling Predicts Tumor Response in Patients with Stage IIIB or IV Non-Small-Cell Lung Cancer.PLoS One. 2015 Jun 5;10(6):e0128970. doi: 10.1371/journal.pone.0128970. eCollection 2015. PLoS One. 2015. PMID: 26047516 Free PMC article.
-
[Preliminary study of MALDI-TOF mass spectrometry-based screening of patients with the NSCLC serum-specific peptides].Zhongguo Fei Ai Za Zhi. 2013 May;16(5):233-9. doi: 10.3779/j.issn.1009-3419.2013.05.04. Zhongguo Fei Ai Za Zhi. 2013. PMID: 23676979 Free PMC article. Chinese.
-
MALDI-TOF-MS analysis in low molecular weight serum peptidome biomarkers for NSCLC.J Clin Lab Anal. 2022 Apr;36(4):e24254. doi: 10.1002/jcla.24254. Epub 2022 Feb 25. J Clin Lab Anal. 2022. PMID: 35212031 Free PMC article.
-
The MALDI-TOF mass spectrometric view of the plasma proteome and peptidome.Clin Chem. 2006 Jul;52(7):1223-37. doi: 10.1373/clinchem.2006.069252. Epub 2006 Apr 27. Clin Chem. 2006. PMID: 16644871 Review.
-
Advances in proteomic strategies toward the early detection of lung cancer.Proc Am Thorac Soc. 2011 May;8(2):183-8. doi: 10.1513/pats.201012-069MS. Proc Am Thorac Soc. 2011. PMID: 21543799 Free PMC article. Review.
Cited by
-
Evaluation of protease inhibitors containing tubes for MS-based plasma peptide profiling studies.J Clin Lab Anal. 2014 Sep;28(5):364-7. doi: 10.1002/jcla.21694. Epub 2014 Mar 19. J Clin Lab Anal. 2014. PMID: 24648264 Free PMC article.
-
Classification of Epidermal Growth Factor Receptor Gene Mutation Status Using Serum Proteomic Profiling Predicts Tumor Response in Patients with Stage IIIB or IV Non-Small-Cell Lung Cancer.PLoS One. 2015 Jun 5;10(6):e0128970. doi: 10.1371/journal.pone.0128970. eCollection 2015. PLoS One. 2015. PMID: 26047516 Free PMC article.
-
Identification of peptide regions of SERPINA1 and ENOSF1 and their protein expression as potential serum biomarkers for gastric cancer.Tumour Biol. 2015 Jul;36(7):5109-18. doi: 10.1007/s13277-015-3163-2. Epub 2015 Feb 14. Tumour Biol. 2015. PMID: 25677901
-
[Detection of Serum Peptides in Patients with Lung Squamous Cell Carcinoma by MALDI-TOF-MS and Analysis of Their Correlation with Chemotherapy Efficacy].Zhongguo Fei Ai Za Zhi. 2017 May 20;20(5):318-325. doi: 10.3779/j.issn.1009-3419.2017.05.04. Zhongguo Fei Ai Za Zhi. 2017. PMID: 28532539 Free PMC article. Clinical Trial. Chinese.
-
Serum peptidome patterns of hepatocellular carcinoma based on magnetic bead separation and mass spectrometry analysis.Diagn Pathol. 2013 Aug 5;8:130. doi: 10.1186/1746-1596-8-130. Diagn Pathol. 2013. PMID: 23915185 Free PMC article.
References
-
- Goldstraw P, Crowley J, Chansky K, et al. International Association for the Study of Lung Cancer International Staging Committee; Participating institutions. The IASLC Lung Cancer Staging Project: Proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM classification of malignant tumours. J Thorac Oncol 2007;2:706–714. - PubMed
-
- Vivian B, Stefan H, Benjamin N, et al. Relevance of circulating biomarkersfor the therapy monitoring and follow‐up investigations in patients with non‐small cell lung cancer. Cancer Biomark 2010;6:191–196. - PubMed
-
- Patz EF, Campa MJ, Gottlin EB, et al. Panel of serum biomarkers for the diagnosis of lung cancer. J Clin Oncol 2007;25:5578–5583. - PubMed
-
- Tarro G, Perna A, Esposito C. Early diagnosis of lung cancer by detection of tumor liberated protein. J Cell Physiol 2005;203:1–5. - PubMed
-
- Sung HJ, Cho JY. Biomarkers for the lung cancer diagnosis and their advances in proteomics. BMB Rep 2008;41:615–625. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

