In vitro loading of human cohesin on DNA by the human Scc2-Scc4 loader complex
- PMID: 22628566
- PMCID: PMC3386075
- DOI: 10.1073/pnas.1206840109
In vitro loading of human cohesin on DNA by the human Scc2-Scc4 loader complex
Abstract
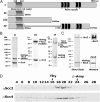
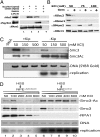
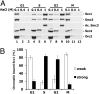
The loading of cohesin onto chromatin requires the heterodimeric complex sister chromatid cohesion (Scc)2 and Scc4 (Scc2/4), which is highly conserved in all species. Here, we describe the purification of the human (h)-Scc2/4 and show that it interacts with h-cohesin and the heterodimeric Smc1-Smc3 complex but not with the Smc1 or Smc3 subunit alone. We demonstrate that both h-Scc2/4 and h-cohesin are loaded onto dsDNA containing the prereplication complex (pre-RC) generated in vitro by Xenopus high-speed soluble extracts. The addition of geminin, which blocks pre-RC formation, prevents the loading of Scc2/4 and cohesin. Xenopus extracts depleted of endogenous Scc2/4 with specific antibodies, although able to form pre-RCs, did not support cohesin loading unless supplemented with purified h-Scc2/4. The results presented here indicate that the Xenopus or h-Scc2/4 complex supports the loading of Xenopus and/or h-cohesin onto pre-RCs formed by Xenopus high-speed extracts. We show that cohesin loaded onto pre-RCs either by h-Scc2/4 and/or the Xenopus complex was dissociated from chromatin by low salt extraction, similar to cohesin loaded onto chromatin in G(1) by HeLa cells in vivo. Replication of cohesin-loaded DNA, both in vitro and in vivo, markedly increased the stability of cohesin associated with DNA. Collectively, these in vitro findings partly recapitulate the in vivo pathway by which sister chromatids are linked together, leading to cohesion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol. 2008;24:105–129. - PubMed
-
- Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. - PubMed
-
- Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. - PubMed
-
- Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat Cell Biol. 2004;6:991–996. - PubMed
-
- Nasmyth K. Cohesin: a catenase with separate entry and exit gates? Nat Cell Biol. 2011;13:1170–1177. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

