Gene regulatory networks in cardiac conduction system development
- PMID: 22628576
- PMCID: PMC4113246
- DOI: 10.1161/CIRCRESAHA.111.260026
Gene regulatory networks in cardiac conduction system development
Abstract
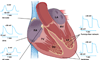
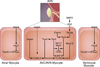
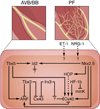
The cardiac conduction system is a specialized tract of myocardial cells responsible for maintaining normal cardiac rhythm. Given its critical role in coordinating cardiac performance, a detailed analysis of the molecular mechanisms underlying conduction system formation should inform our understanding of arrhythmia pathophysiology and affect the development of novel therapeutic strategies. Historically, the ability to distinguish cells of the conduction system from neighboring working myocytes presented a major technical challenge for performing comprehensive mechanistic studies. Early lineage tracing experiments suggested that conduction cells derive from cardiomyocyte precursors, and these claims have been substantiated by using more contemporary approaches. However, regional specialization of conduction cells adds an additional layer of complexity to this system, and it appears that different components of the conduction system utilize unique modes of developmental formation. The identification of numerous transcription factors and their downstream target genes involved in regional differentiation of the conduction system has provided insight into how lineage commitment is achieved. Furthermore, by adopting cutting-edge genetic techniques in combination with sophisticated phenotyping capabilities, investigators have made substantial progress in delineating the regulatory networks that orchestrate conduction system formation and their role in cardiac rhythm and physiology. This review describes the connectivity of these gene regulatory networks in cardiac conduction system development and discusses how they provide a foundation for understanding normal and pathological human cardiac rhythms.
Figures
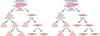

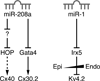
References
-
- Echt DS, Liebson PR, Mitchell LB, et al. Mortality and morbidity in patients receiving encainide, flecainide, or placebo: the cardiac arrhythmia suppression trial. N Engl J Med. 1991;324:781–788. - PubMed
-
- Splawski I, Tristani-Firouzi M, Lehmann MH, Sanguinetti MC, Keating MT. Mutations in the hmink gene cause long QT syndrome and suppress Iks function. Nat Genet. 1997;17:338–340. - PubMed
-
- Keating MT, Sanguinetti MC. Pathophysiology of ion channel mutations. Curr Opin Genet Dev. 1996;6:326–333. - PubMed
-
- Keating MT, Sanguinetti MC. Molecular genetic insights into cardiovascular disease. Science. 1996;272:681–685. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

