Glycosylphosphatidylinositol anchors regulate glycosphingolipid levels
- PMID: 22628614
- PMCID: PMC3540855
- DOI: 10.1194/jlr.M025692
Glycosylphosphatidylinositol anchors regulate glycosphingolipid levels
Abstract
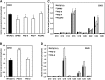
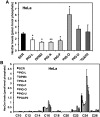
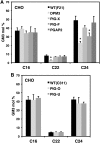
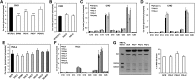
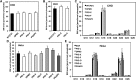
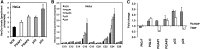
Glycosylphosphatidylinositol (GPI) anchor biosynthesis takes place in the endoplasmic reticulum (ER). After protein attachment, the GPI anchor is transported to the Golgi where it undergoes fatty acid remodeling. The ER exit of GPI-anchored proteins is controlled by glycan remodeling and p24 complexes act as cargo receptors for GPI anchor sorting into COPII vesicles. In this study, we have characterized the lipid profile of mammalian cell lines that have a defect in GPI anchor biosynthesis. Depending on which step of GPI anchor biosynthesis the cells were defective, we observed sphingolipid changes predominantly for very long chain monoglycosylated ceramides (HexCer). We found that the structure of the GPI anchor plays an important role in the control of HexCer levels. GPI anchor-deficient cells that generate short truncated GPI anchor intermediates showed a decrease in very long chain HexCer levels. Cells that synthesize GPI anchors but have a defect in GPI anchor remodeling in the ER have a general increase in HexCer levels. GPI-transamidase-deficient cells that produce no GPI-anchored proteins but generate complete free GPI anchors had unchanged levels of HexCer. In contrast, sphingomyelin levels were mostly unaffected. We therefore propose a model in which the transport of very long chain ceramide from the ER to Golgi is regulated by the transport of GPI anchor molecules.
Figures



References
-
- Maeda Y., Ashida H., Kinoshita T. 2006. CHO glycosylation mutants: GPI anchor. Methods Enzymol. 416: 182–205 - PubMed
-
- Bütikofer P., Malherbe T., Boschung M., Roditi I. 2001. GPI-anchored proteins: now you see ‘em, now you don't. FASEB J. 15: 545–548 - PubMed
-
- Tanaka S., Maeda Y., Tashima Y., Kinoshita T. 2004. Inositol deacylation of glycosylphosphatidylinositol-anchored proteins is mediated by mammalian PGAP1 and yeast Bst1p. J. Biol. Chem. 279: 14256–14263 - PubMed
-
- Fujita M., Maeda Y., Ra M., Yamaguchi Y., Taguchi R., Kinoshita T. 2009. GPI glycan remodeling by PGAP5 regulates transport of GPI-anchored proteins from the ER to the Golgi. Cell. 139: 352–365 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

