Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation
- PMID: 22628656
- PMCID: PMC3526189
- DOI: 10.1126/science.1218595
Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation
Abstract
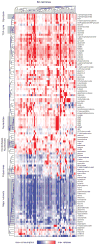
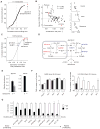
Metabolic reprogramming has been proposed to be a hallmark of cancer, yet a systematic characterization of the metabolic pathways active in transformed cells is currently lacking. Using mass spectrometry, we measured the consumption and release (CORE) profiles of 219 metabolites from media across the NCI-60 cancer cell lines, and integrated these data with a preexisting atlas of gene expression. This analysis identified glycine consumption and expression of the mitochondrial glycine biosynthetic pathway as strongly correlated with rates of proliferation across cancer cells. Antagonizing glycine uptake and its mitochondrial biosynthesis preferentially impaired rapidly proliferating cells. Moreover, higher expression of this pathway was associated with greater mortality in breast cancer patients. Increased reliance on glycine may represent a metabolic vulnerability for selectively targeting rapid cancer cell proliferation.
Conflict of interest statement
All authors declare no competing financial interests.
Figures



Comment in
-
Cancer. Systems biology, metabolomics, and cancer metabolism.Science. 2012 May 25;336(6084):990-1. doi: 10.1126/science.1223066. Science. 2012. PMID: 22628644 No abstract available.
-
Metabolism: craving for glycine.Nat Rev Clin Oncol. 2012 Jun 12;9(8):430. doi: 10.1038/nrclinonc.2012.103. Nat Rev Clin Oncol. 2012. PMID: 22688577 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

