Assessment method for a power analysis to identify differentially expressed pathways
- PMID: 22629411
- PMCID: PMC3356338
- DOI: 10.1371/journal.pone.0037510
Assessment method for a power analysis to identify differentially expressed pathways
Abstract
Gene expression data can provide a very rich source of information for elucidating the biological function on the pathway level if the experimental design considers the needs of the statistical analysis methods. The purpose of this paper is to provide a comparative analysis of statistical methods for detecting the differentially expression of pathways (DEP). In contrast to many other studies conducted so far, we use three novel simulation types, producing a more realistic correlation structure than previous simulation methods. This includes also the generation of surrogate data from two large-scale microarray experiments from prostate cancer and ALL. As a result from our comprehensive analysis of 41,004 parameter configurations, we find that each method should only be applied if certain conditions of the data from a pathway are met. Further, we provide method-specific estimates for the optimal sample size for microarray experiments aiming to identify DEP in order to avoid an underpowered design. Our study highlights the sensitivity of the studied methods on the parameters of the system.
Conflict of interest statement
Figures
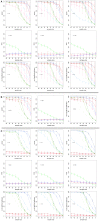
 (light color),
(light color),  (medium color),
(medium color),  (dark color).
(dark color).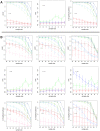
 (light color),
(light color),  (medium color),
(medium color),  (dark color). Simulated data are from the protein network of yeast .
(dark color). Simulated data are from the protein network of yeast .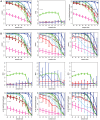
 (light color),
(light color),  (medium color),
(medium color),  (dark color). Simulated data are from the transcriptional regulatory network of yeast .
(dark color). Simulated data are from the transcriptional regulatory network of yeast .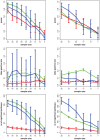

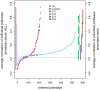
 ), ST II (orange -
), ST II (orange -  ), ST III (purple,
), ST III (purple,  ) and ST IV (brown -
) and ST IV (brown -  ) the projections of the range of correlation values is shown on the right-hand side.
) the projections of the range of correlation values is shown on the right-hand side.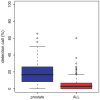
References
-
- Alon U. An Introduction to Systems Biology: Design Principles of Biological Circuits. Boca Raton, FL: Chapman & Hall/CRC; 2006.
-
- Emmert-Streib F, Dehmer M, editors. Medical Biostatistics for Complex Diseases. Weinheim: Wiley-Blackwell; 2010.
-
- Kauffman S. Metabolic stability and epigenesis in randomly constructed genetic nets. Journal of Theoretical Biology. 1969;22:437–467. - PubMed
-
- Niiranen S, Ribeiro A, editors. Information Processing and Biological Systems. Berlin: Springer; 2011.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

