Monitoring lipid anchor organization in cell membranes by PIE-FCCS
- PMID: 22631607
- PMCID: PMC3626236
- DOI: 10.1021/ja300374c
Monitoring lipid anchor organization in cell membranes by PIE-FCCS
Abstract
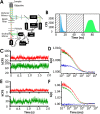
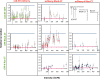
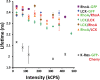
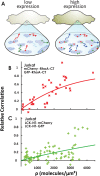
This study examines the dynamic co-localization of lipid-anchored fluorescent proteins in living cells using pulsed-interleaved excitation fluorescence cross-correlation spectroscopy (PIE-FCCS) and fluorescence lifetime analysis. Specifically, we look at the pairwise co-localization of anchors from lymphocyte cell kinase (LCK: myristoyl, palmitoyl, palmitoyl), RhoA (geranylgeranyl), and K-Ras (farnesyl) proteins in different cell types. In Jurkat cells, a density-dependent increase in cross-correlation among RhoA anchors is observed, while LCK anchors exhibit a more moderate increase and broader distribution. No correlation was detected among K-Ras anchors or between any of the different anchor types studied. Fluorescence lifetime data reveal no significant Förster resonance energy transfer in any of the data. In COS 7 cells, minimal correlation was detected among LCK or RhoA anchors. Taken together, these observations suggest that some lipid anchors take part in anchor-specific co-clustering with other existing clusters of native proteins and lipids in the membrane. Importantly, these observations do not support a simple interpretation of lipid anchor-mediated organization driven by partitioning based on binary lipid phase separation.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

