Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair
- PMID: 22632967
- PMCID: PMC3616325
- DOI: 10.1016/j.cell.2012.03.043
Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair
Abstract
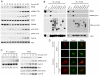
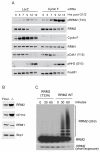
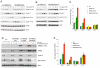
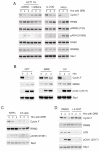
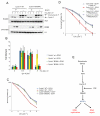
F-box proteins are the substrate binding subunits of SCF (Skp1-Cul1-F-box protein) ubiquitin ligase complexes. Using affinity purifications and mass spectrometry, we identified RRM2 (the ribonucleotide reductase family member 2) as an interactor of the F-box protein cyclin F. Ribonucleotide reductase (RNR) catalyzes the conversion of ribonucleotides to deoxyribonucleotides (dNTPs), which are necessary for both replicative and repair DNA synthesis. We found that, during G2, following CDK-mediated phosphorylation of Thr33, RRM2 is degraded via SCF(cyclin F) to maintain balanced dNTP pools and genome stability. After DNA damage, cyclin F is downregulated in an ATR-dependent manner to allow accumulation of RRM2. Defective elimination of cyclin F delays DNA repair and sensitizes cells to DNA damage, a phenotype that is reverted by expressing a nondegradable RRM2 mutant. In summary, we have identified a biochemical pathway that controls the abundance of dNTPs and ensures efficient DNA repair in response to genotoxic stress.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures

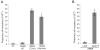
References
-
- Andersson B, Hou SM, Lambert B. Mutations causing defective splicing in the human hprt gene. Environmental and molecular mutagenesis. 1992;20:89–95. - PubMed
-
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

