Identification of the cortical neurons that mediate antidepressant responses
- PMID: 22632977
- PMCID: PMC3397430
- DOI: 10.1016/j.cell.2012.03.038
Identification of the cortical neurons that mediate antidepressant responses
Abstract
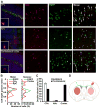
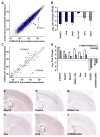
Our understanding of current treatments for depression, and the development of more specific therapies, is limited by the complexity of the circuits controlling mood and the distributed actions of antidepressants. Although the therapeutic efficacy of serotonin-specific reuptake inhibitors (SSRIs) is correlated with increases in cortical activity, the cell types crucial for their action remain unknown. Here we employ bacTRAP translational profiling to show that layer 5 corticostriatal pyramidal cells expressing p11 (S100a10) are strongly and specifically responsive to chronic antidepressant treatment. This response requires p11 and includes the specific induction of Htr4 expression. Cortex-specific deletion of p11 abolishes behavioral responses to SSRIs, but does not lead to increased depression-like behaviors. Our data identify corticostriatal projection neurons as critical for the response to antidepressants, and suggest that the regulation of serotonergic tone in this single cell type plays a pivotal role in antidepressant therapy.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures




References
-
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. - PubMed
-
- Covington HE, 3rd, Lobo MK, Maze I, Vialou V, Hyman JM, Zaman S, LaPlant Q, Mouzon E, Ghose S, Tamminga CA, et al. Antidepressant effect of optogenetic stimulation of the medial prefrontal cortex. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2010;30:16082–16090. - PMC - PubMed
-
- Cryan JF, Mombereau C, Vassout A. The tail suspension test as a model for assessing antidepressant activity: review of pharmacological and genetic studies in mice. Neurosci Biobehav Rev. 2005;29:571–625. - PubMed
-
- Delgado MR. Reward-related responses in the human striatum. Ann N Y Acad Sci. 2007;1104:70–88. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

