Antibiotic-induced bacterial cell death exhibits physiological and biochemical hallmarks of apoptosis
- PMID: 22633370
- PMCID: PMC3710583
- DOI: 10.1016/j.molcel.2012.04.027
Antibiotic-induced bacterial cell death exhibits physiological and biochemical hallmarks of apoptosis
Abstract
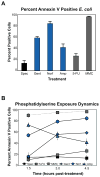
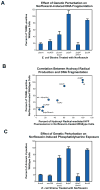
Programmed cell death is a gene-directed process involved in the development and homeostasis of multicellular organisms. The most common mode of programmed cell death is apoptosis, which is characterized by a stereotypical set of biochemical and morphological hallmarks. Here we report that Escherichia coli also exhibit characteristic markers of apoptosis-including phosphatidylserine exposure, chromosome condensation, and DNA fragmentation-when faced with cell death-triggering stress, namely bactericidal antibiotic treatment. Notably, we also provide proteomic and genetic evidence for the ability of multifunctional RecA to bind peptide sequences that serve as substrates for eukaryotic caspases, and regulation of this phenotype by the protease, ClpXP, under conditions of cell death. Our findings illustrate that prokaryotic organisms possess mechanisms to dismantle and mark dying cells in response to diverse noxious stimuli and suggest that elaborate, multilayered proteolytic regulation of these features may have evolved in eukaryotes to harness and exploit their deadly potential.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures




Comment in
-
When death was young: an ancestral apoptotic network in bacteria.Mol Cell. 2012 Jun 8;46(5):552-4. doi: 10.1016/j.molcel.2012.05.032. Mol Cell. 2012. PMID: 22681882
References
-
- Ameisen JC. The origin of programmed cell death. Science. 1996;272:1278–1279. - PubMed
-
- Ameisen JC. On the origin, evolution, and nature of programmed cell death: a timeline of four billion years. Cell Death Differ. 2002;9:367–393. - PubMed
-
- Blackstone NW, Green DR. The evolution of a mechanism of cell suicide. Bioessays. 1999;21:84–88. - PubMed
-
- Broker LE, Kruyt FA, Giaccone G. Cell death independent of caspases: a review. Clin Cancer Res. 2005;11:3155–3162. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

